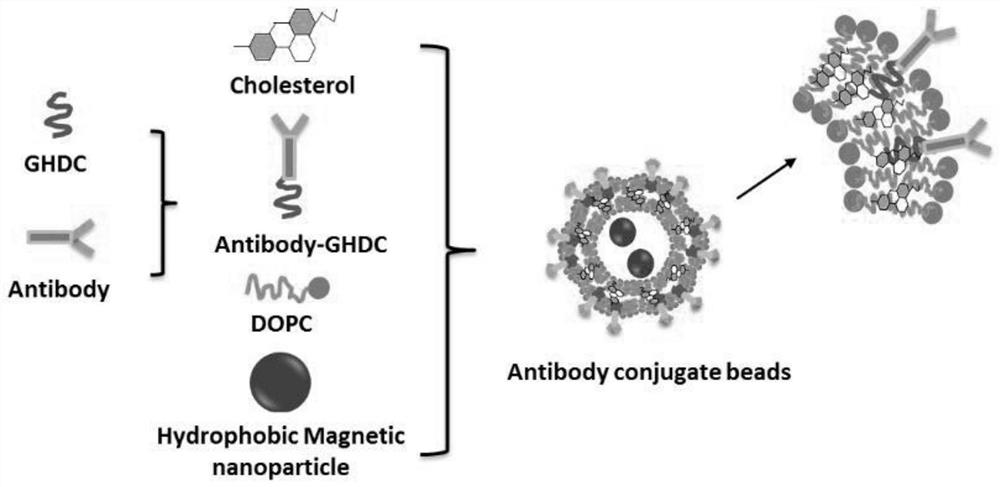
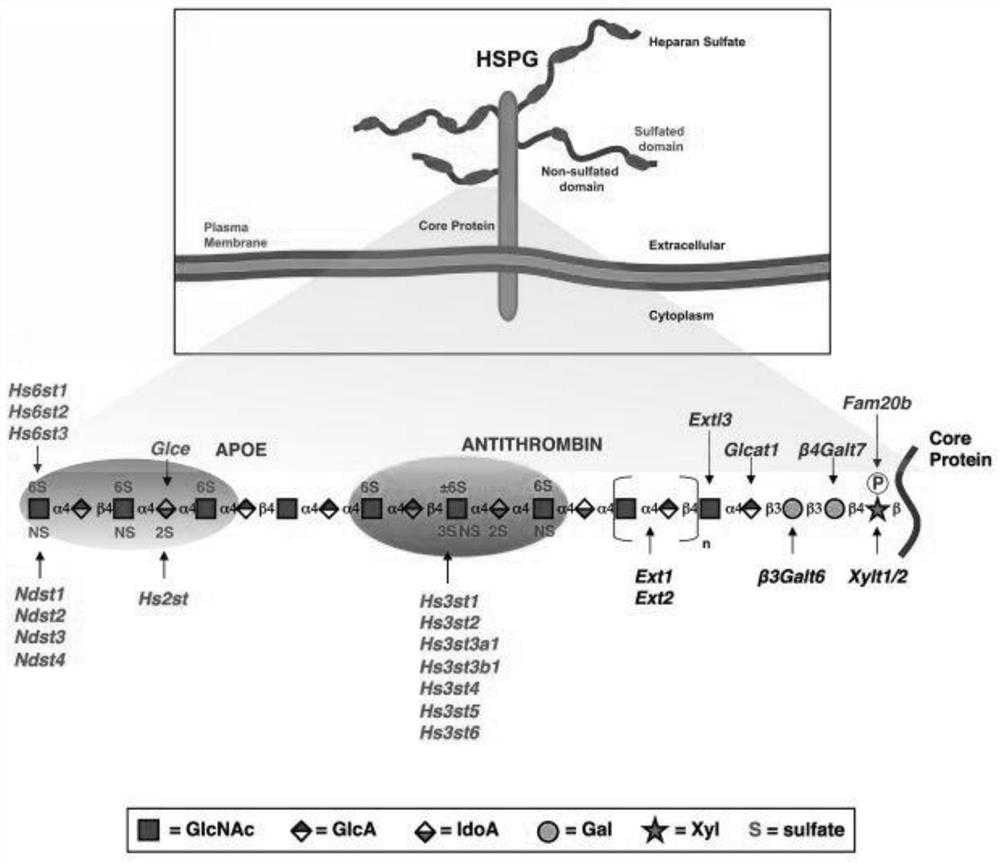
Method for separating and identifying biliary tract cancer peripheral blood CTC (circulating tumor cells) and detecting downstream genes by HSPG (high-throughput sequencing assay)
A technology of peripheral blood and biliary tract, applied in the field of biological and medical detection, can solve the problems of short survival period, low degree of cell differentiation, poor response to chemotherapy, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] A method for separating and identifying CTCs in peripheral blood of biliary tract cancer by HSPG, comprising the steps of:
[0082] (1) Sampling: Take 7.5 mL of peripheral blood from patients with biliary tract cancer, place it in an anticoagulant tube and mix the whole blood sample;
[0083] (2) Remove plasma protein and plasma (clear) nucleic acid from the blood sample collected in step (1), add buffer solution to the whole blood, remove the supernatant by layered centrifugation, and gently shake the centrifuge tube to mix and precipitate the cells;
[0084] (3) Removal of red blood cells: After mixing, add the lysate to the centrifuge tube, place the centrifuge tube in a vertical mixer to mix, centrifuge to discard the supernatant, shake the centrifuge tube to mix and settle the cells, and then add buffer;
[0085] (4) Layered centrifugation: Add the layered liquid into a new centrifuge tube, superimpose all the liquid in step (3) on the top layer of the layered liqu...
Embodiment 2
[0122] A HSPG method for downstream gene detection of peripheral blood CTCs of biliary tract cancer, using a high-throughput sequencing method to perform genome-wide amplification of nucleic acids derived from blood circulating tumor cells of biliary tract cancer, the specific steps are as follows:
[0123] (1) Thaw and lyse the frozen CTC PCR plate on ice, and add 0.4M HCl to each well after lysing;
[0124] (2) Multiple replacement amplification was used for whole genome amplification, and the reaction mixture was prepared. For each reaction, sterilized water, 10× reaction buffer, BSA, DTT, dNTPs in the replication genome amplification kit, random six After the aggregates are mixed and acted according to the amount, they are then incubated on a thermal cycler at 30°C;
[0125] (3) The paramagnetic beads in the nucleic acid purification kit are used to terminate the subsequent MDA reaction, add 100 μL of the paramagnetic beads in the nucleic acid purification kit to each reac...
Embodiment 3
[0160] (1) Determine the sample size:
[0161] (a) 100 patients with biliary tract cancer were enrolled in the early stage, the detection system and indicators were evaluated, and the follow-up sample size was calculated.
[0162] (b) According to the previous research results, statisticians will preliminarily calculate and count the number of CTCs identified by HSPG antibody sorting specific for biliary tract cancer, and compare the sorting efficiency and differences among individual individuals, and count the sensitivity and specificity of markers , to further calculate the total sample size required, it is expected to enroll about 500 patients.
[0163] (c) According to the previous research results, the number and frequency of gene mutations derived from CTC nucleic acid are further counted, and the total sample size required is calculated. It is estimated that about 500 patients will be enrolled.
[0164] (2) Inclusion criteria:
[0165] The subjects of the study were p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com