Escherichia coli integrated with amino acid oxidase and glucose dehydrogenase
A technology of glucose dehydrogenase and Escherichia coli, applied in the field of bioengineering, can solve the problems of increasing the complexity of antibiotics and inducers, and increasing the production cost of inducers, achieving strong environmental adaptability, reducing operation steps, and good application characteristics. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1: Screening of Chromosomal Integration Sites
[0033] 1. The chromosome of Escherichia coli BL21 (DE3) with a total sequence length of 4.6Mbp is divided into six regions, and two sites are selected in each region to integrate L-amino acid oxidase and glucose dehydrogenase, and then The strains with L-amino acid oxidase and glucose dehydrogenase integrated at each site were tested for activity, so as to determine the best integration site. The grouped regions and sites are 0~0.76Mbp(dkgB,eaeH), 0.76~1.53Mbp(lysO,bluF), 1.53~2.28Mbp(tam,glsB), 2.28~3.04Mbp(rcs,mntH), 3.04 ~3.8Mbp(nupG, pitB), 3.8~4.6Mbp(rbsA, pgi).
[0034] (1) Extraction of plasmid
[0035] Plasmid extraction was performed according to the instructions on the plasmid mini-extraction kit of Shanghai Jierui Company.
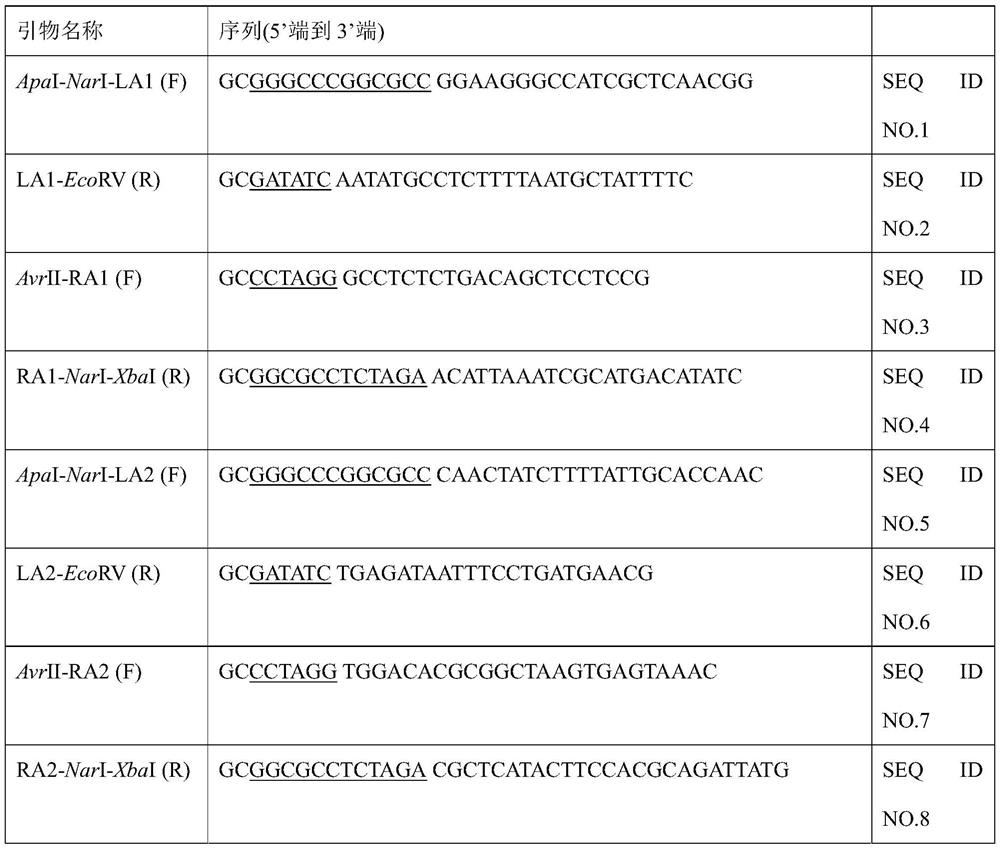
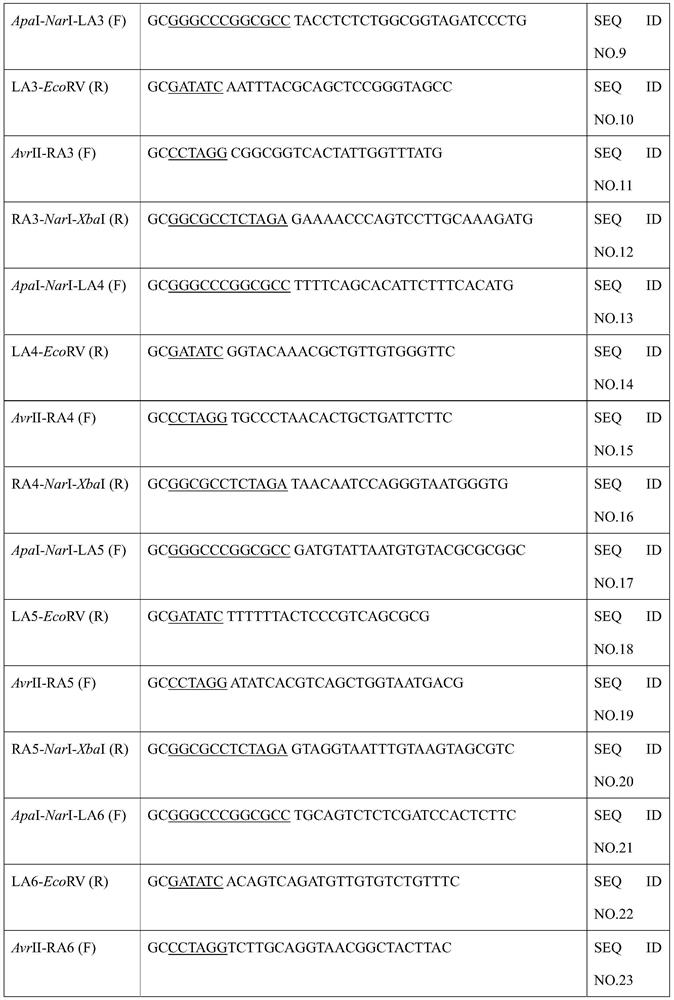
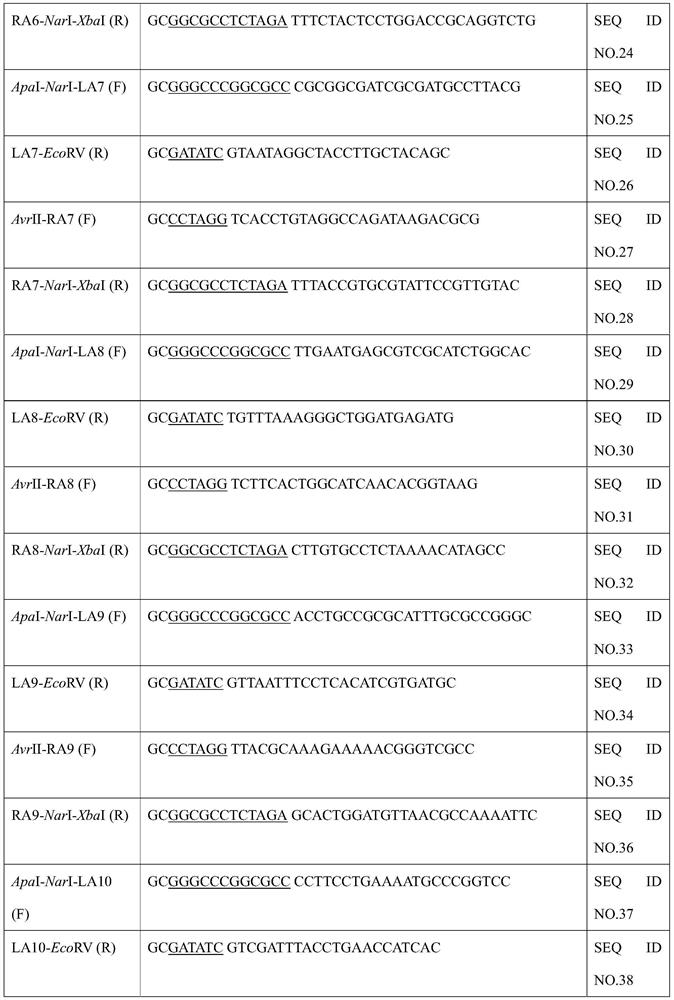
[0036] (2) Construction of pIn-LA-cat-RA plasmid
[0037] The primers used during the construction are listed in Table 1. Chloramphenicol resistance gene was amplified from pKD3...
Embodiment 2
[0076] This example integrates different combinations of L-amino acid oxidase (pmaao, cmaao) and glucose dehydrogenase (gsgdh, hmgdh, bsgdh) at the optimal integration site nupG screened, and inoculates the integrated strains of different combinations in Cultivate overnight in the test tube, then inoculate 50 mL of fresh LB medium with 1% inoculum, put it into a vibrating shaker, and cultivate at 25°C and 200 rpm for 24 hours. After 24 hours, the cells were collected by centrifugation at 8000 rpm for 10 min at 4°C. Using 50mM sodium dihydrogen phosphate-disodium hydrogen phosphate solution at pH 7.5 as the reaction buffer, the whole-cell catalytic reaction system is: 50g / L wet cells, 35°C, 3g / L L-phenylalanine, transformed The time is 5h. After conversion, the yield of phenylpyruvate was determined by high performance liquid chromatography (HPLC). Take 20mL, OD 600 3.5 bacteria solution, centrifuged at 8000 rpm for 10 min at 4°C, and then collected the cells. Use 20mL, 35m...
Embodiment 3
[0082] This example integrates multiple copies of L-amino acid oxidase pmaao and glucose dehydrogenase bsgdh at the screened optimal integration sites glsB, nupG, and pitB, and tests the multi-site multi-copy integration of pmaao and bsgdh to transform the strain into L- Yield of phenylalanine to phenylpyruvate and activity of glucose dehydrogenase.
[0083] Genes with different copy numbers were sequentially integrated at the sites screened in Example 1, and multi-copy integration followed the FLP recombination method (rapid and reliable strategy for chromosomal integration of gene(s) with multiple copies. Scientific Reports 2015, 5.). Inoculate E.coli BL21 strains with different copy numbers of pmaao and bsgdh into test tubes for overnight culture, then inoculate 1% of the inoculum into 50 mL of fresh LB medium, put it into a shaking shaker, and incubate at 25°C 1. Cultivate for 24 hours under the condition of 200 rpm. After 24 hours, the cells were collected by centrifugat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com