Method for detecting huanglongbing bacteria based on aPCR and DNA walker
A detection method, Huanglong's technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of "sequence identification, low reliability of detection results, expensive detection costs, etc., to reduce detection costs, Low-cost, high-accuracy results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0051] Embodiments of the present invention will be described in further detail below in conjunction with the accompanying drawings.
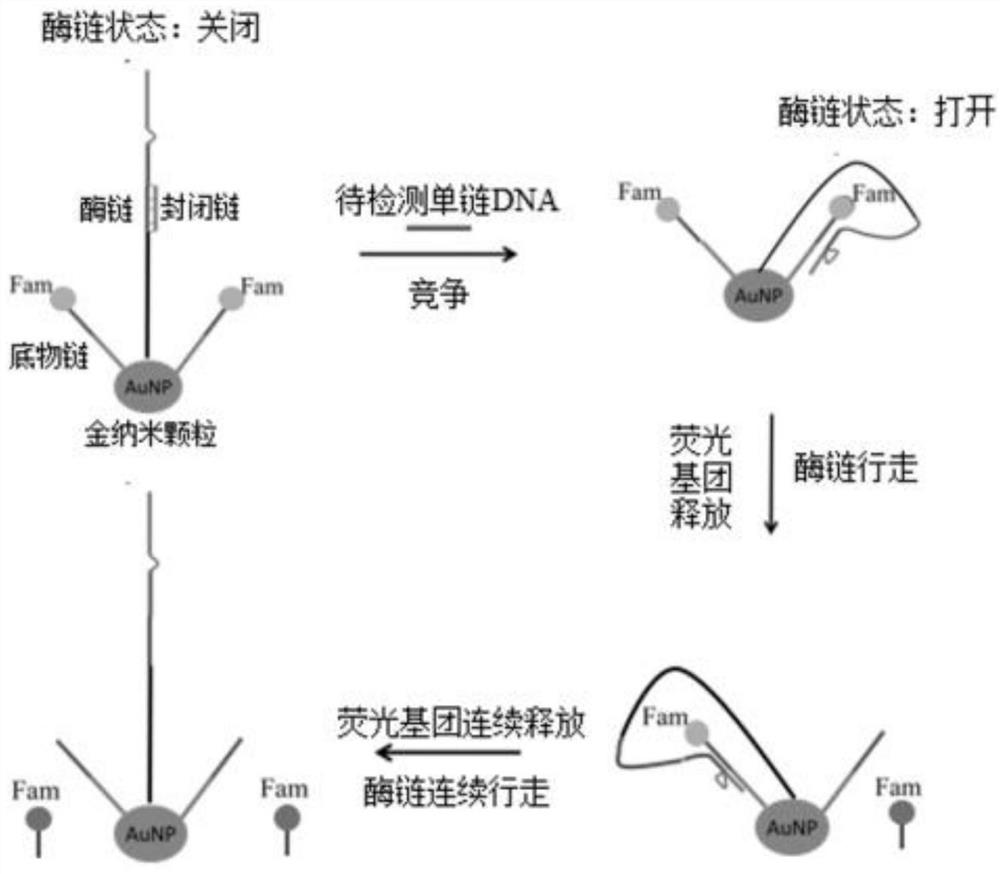
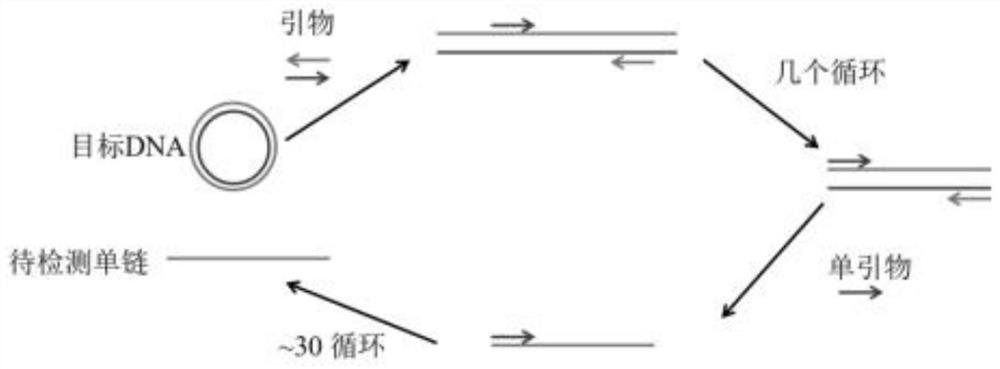
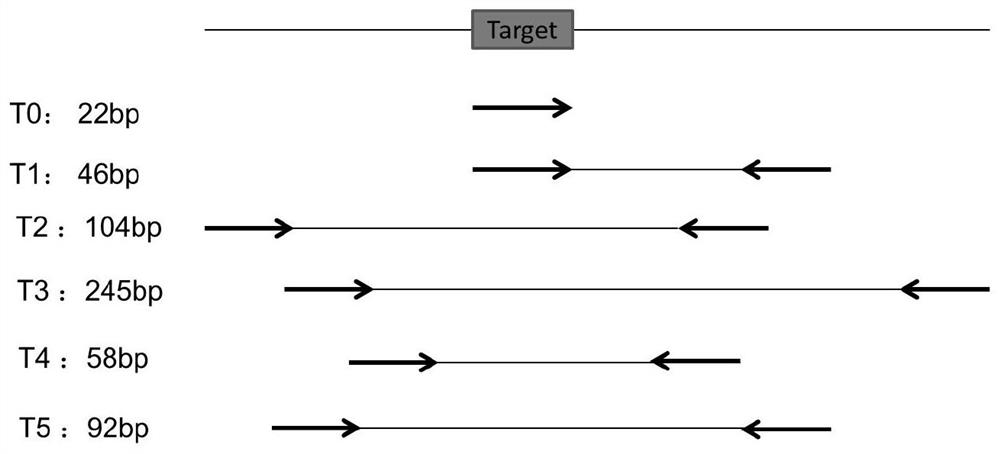
[0052] see figure 1 As shown, the embodiment of the present invention provides a method for detecting Huanglong bacteria based on aPCR and DNA walker. The present invention selects the single-stranded DNA fragments amplified by aPCR (asymmetric PCR, asymmetric amplification) related genes of Huanglong bacteria as the to-be-detected The target, coupled with the DNA Walker detection system, can detect a large number of samples in a short period of time, the cost of high-throughput detection is low, and the detection cycle is short (Sanger sequencing generally takes 8 hours, including sample processing, dideoxy amplification, on-board detection, etc., and this technology can be completed within 3 hours, shortening the time by 63%); at the same time, use DNAzyme (Deoxyribozyme, deoxyribozyme, hereinafter referred to as specific enzyme chain DNAzyme...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com