Aptamer design method based on single-nucleotide molecular docking
A nucleic acid aptamer, molecular docking technology, applied in biochemical equipment and methods, genetic engineering, microbial assay/inspection, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
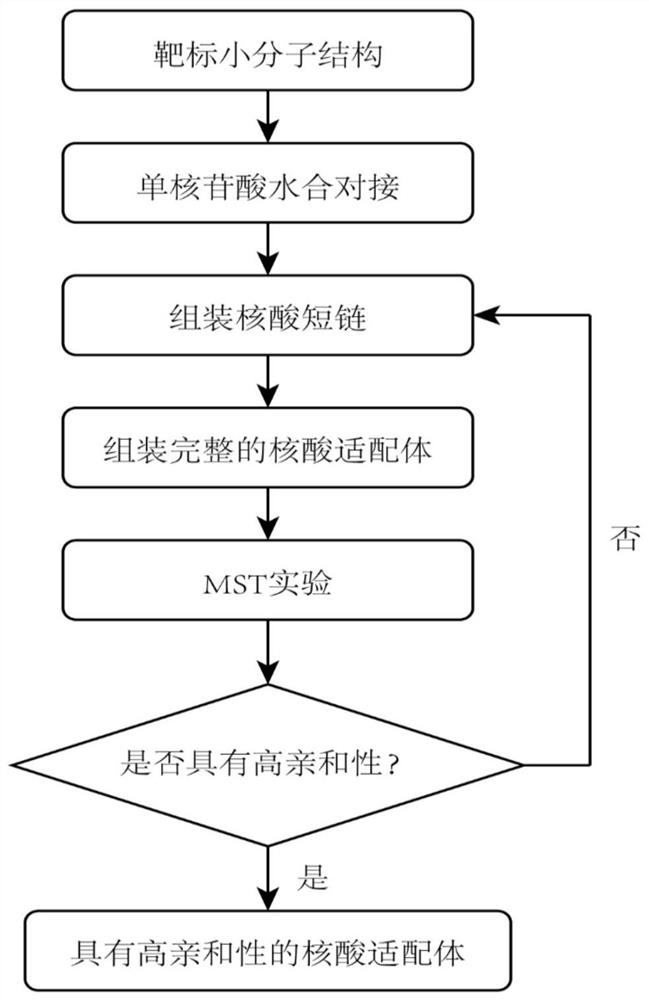
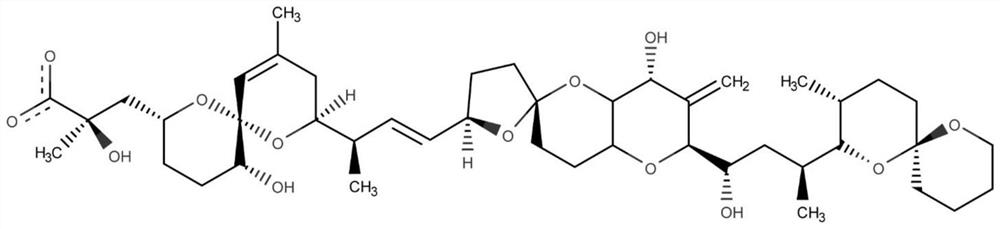
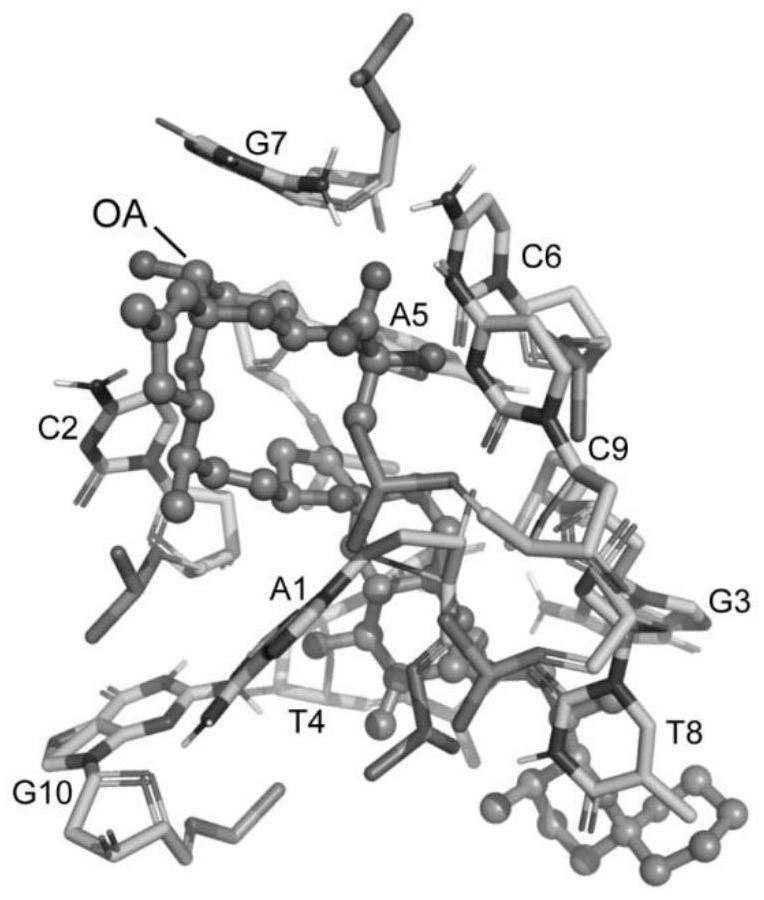
[0034] The specific process of the method of the present invention will be further described below by taking the computational design method of the nucleic acid aptamer OA-D1 targeting OA (Okadaic acid) as an example.
[0035] Step 1: Perform hydration docking of single nucleotides to obtain the position of single nucleotides bound to the surface of small toxin molecules;
[0036] The first step aims to obtain the positions of mononucleotides bound on the OA surface. The molecular docking method can quickly find the main binding site of OA and the binding position of each nucleotide (A, C, G, T) on its surface. In the first docking, the receptor molecule is the toxin small molecule OA, its structure comes from the PDB database (ID: 2I4E), and the ligand molecule is each nucleotide; in the second molecular docking, the receptor molecule is the toxin small molecule OA And the nucleotides retained after docking, the ligand molecule is still each nucleotide. The software used fo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com