Host removing method and kit for metagenomes of pathogen microorganisms
A metagenomic and microbial technology, applied in the field of pathogenic microbial metagenomic dehosting methods and kits, can solve the problems of pathogen reduction, human cell enrichment, etc., and achieve the effect of maintaining integrity and providing detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] The denaturant composition of design experiment group is: the saponin of 5%, Triton X-100 1%, Digitonin2%, CHAPS 1%;
[0042] The denaturant composition of the control group is: Triton X-100 1%, saponin 1%;
[0043] The rest of the experimental conditions were controlled the same.
[0044] experiment procedure:
[0045] Sample preparation:
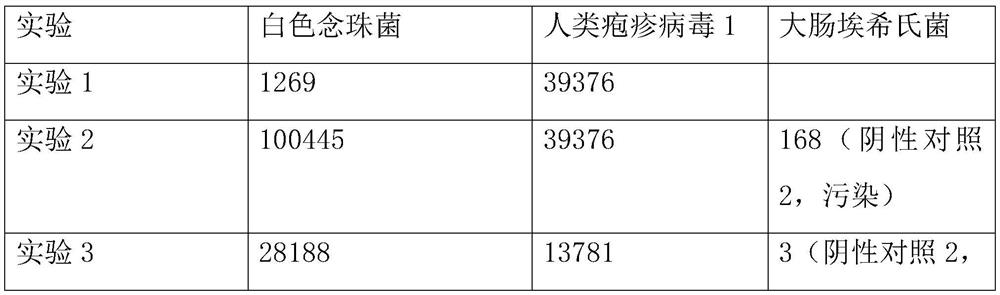
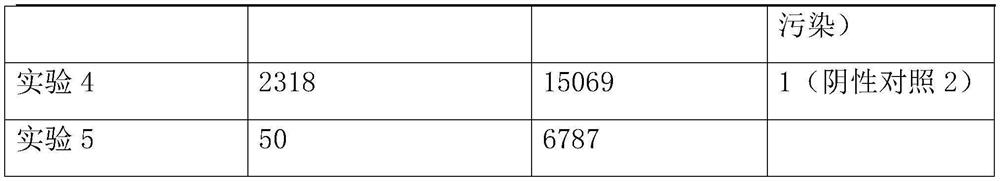
[0046] Take 1.5mL samples of alveolar lavage fluid from 3 patients with clinical infectious diseases, and place them in centrifuge tubes. The patients with clinical infectious diseases were selected from the First Affiliated Hospital of Zhejiang University.
[0047] experiment procedure:
[0048] Take the samples from patients with the same clinical infectious disease and divide them into three equal parts, each containing 1.5mL of samples, and use them as the experimental samples of the experimental group and the control group respectively. A total of 3 experiments were conducted on the same clinical infectious disease patients;...
Embodiment 2
[0055] The denaturant composition of design experiment group 1 is: Triton X-100 0.5%, saponin5%;
[0056] The denaturant components of experimental group 2 are: Triton X-100 0.5%, saponin 1%;
[0057] The denaturant components of experimental group 3 are: Triton X-100 1%, saponin 2%;
[0058] The denaturant composition of experimental group 4 is: Triton X-100 1%, saponin 3%;
[0059] The denaturant composition of experimental group 5 is: Triton X-100 1%, saponin10%;
[0060] The rest of the experimental conditions were controlled the same.
[0061] Sample preparation:
[0062] Take 1.5mL samples of alveolar lavage fluid from 3 patients with clinical infectious diseases, and place them in centrifuge tubes. The patients with clinical infectious diseases were selected from the First Affiliated Hospital of Zhejiang University.
[0063] experiment procedure:
[0064] Samples from patients with the same clinical infectious disease were divided into five equal parts, each contai...
Embodiment 3
[0072] Control group:
[0073] The denaturant composition of design experiment group is: the saponin of 5%, Triton X-100 1%, Digitonin2%, CHAPS 1%;
[0074] The control group does not apply to the human source kit;
[0075] Sample preparation:
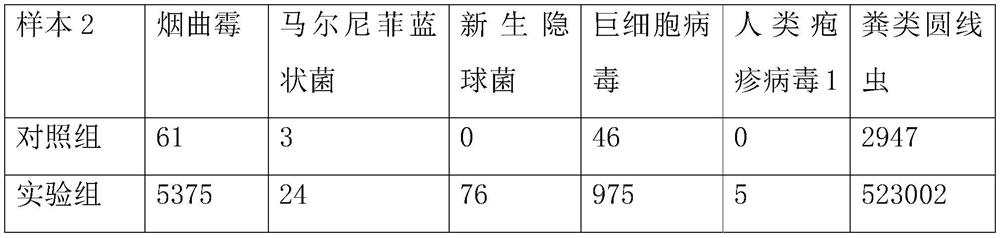
[0076] Take 1.5mL samples of alveolar lavage fluid from multiple patients with clinical infectious diseases and place them in centrifuge tubes. The patients with clinical infectious diseases are selected from the First Affiliated Hospital of Zhejiang University.
[0077] Experiments in the experimental group:
[0078] Take the samples from patients with the same clinical infectious disease and divide them into two equal parts, each containing 1.5mL of samples, and use them as the experimental samples of the experimental group and the control group respectively, and conduct 3 experiments on the same clinical infectious disease patients;
[0079] Experiments in the control group: Directly use Xunminkang Second Generation Sequencing Li...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com