Transcriptome analysis method and system without reference genome sequence
A technology of transcriptome analysis and reference genome, which is applied in the field of transcriptome sequencing data analysis, can solve the problems of transcriptome analysis without reference genome sequence and achieve high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
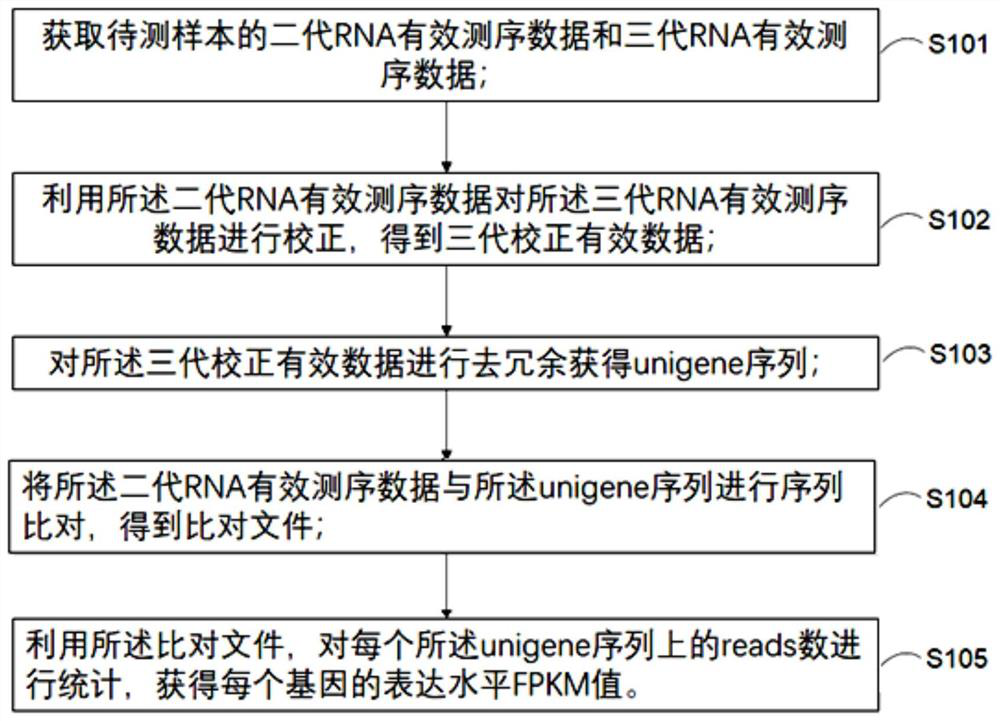
[0047] In this embodiment, a transcriptome analysis method without a reference genome sequence is provided. The flow chart of the analysis method is as follows figure 1 shown, including:
[0048] Step S101, obtaining valid second-generation RNA sequencing data and third-generation RNA effective sequencing data of the sample to be tested;
[0049] Step S102, using the second-generation RNA effective sequencing data to correct the third-generation RNA effective sequencing data to obtain the third-generation corrected effective data;
[0050] Step S103, performing de-redundancy on the three generations of corrected valid data to obtain a unigene sequence;
[0051] Step S104, performing sequence alignment on the second-generation RNA effective sequencing data and the unigene sequence to obtain an alignment file;
[0052] Step S105, using the alignment file, counting the number of reads on each unigene sequence to obtain the expression level FPKM value of each gene.
[0053] The...
Embodiment 2
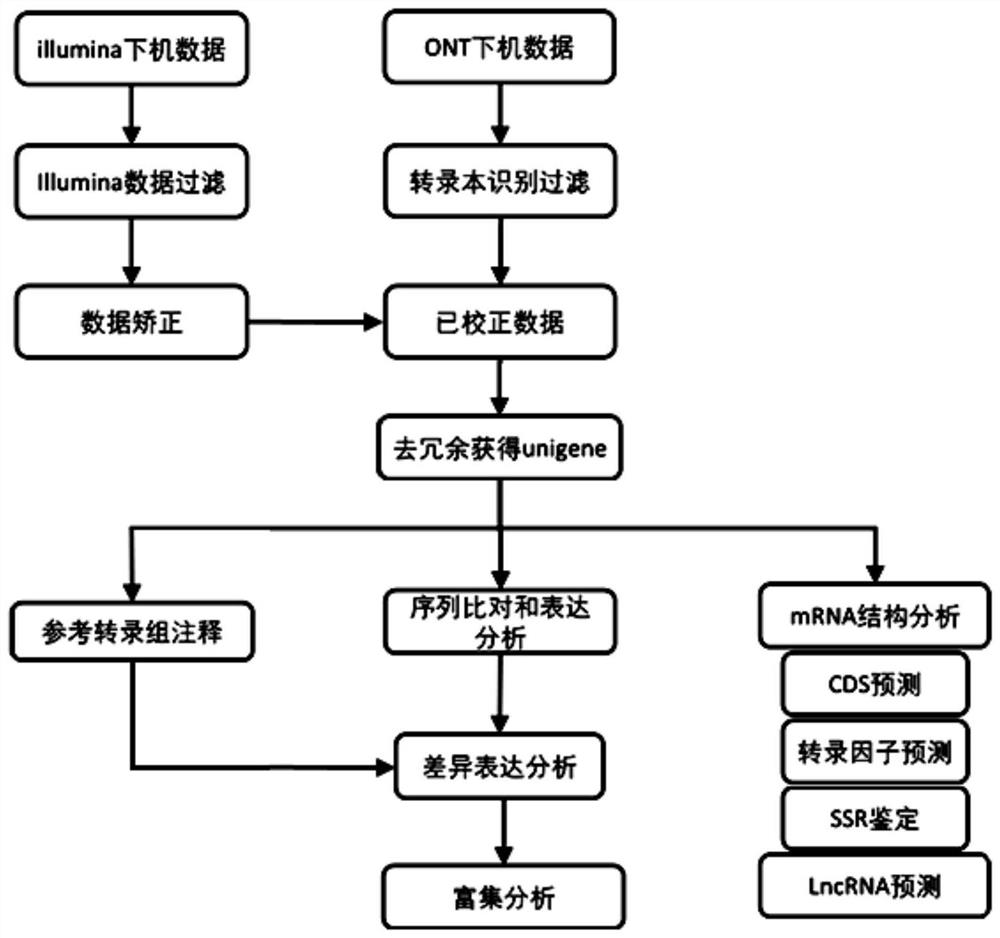
[0067] In this embodiment, a detailed transcriptome analysis method without a reference genome sequence is provided, wherein, figure 2 A detailed schematic flow diagram of the analysis method is shown, specifically including the following steps:
[0068] (1) Extract total RNA from biological samples without a reference genome, build libraries on the illumina platform and ONT sequencing platform, perform sequencing on the machine, and obtain the original sequencing data.
[0069] (2) The off-machine data of the Illumina platform is in fastq format, which can be further analyzed directly. The off-machine data of the ONT platform is in the fast5 format. It is necessary to use the self-developed software guppy v3.6.0 of the ONT platform to use high-precision identification configuration files to basecall the original data and obtain the fastq file.
[0070] (3) For the data on the illumina platform, filter and clean through the software fastp, and filter the data with reads leng...
Embodiment 3
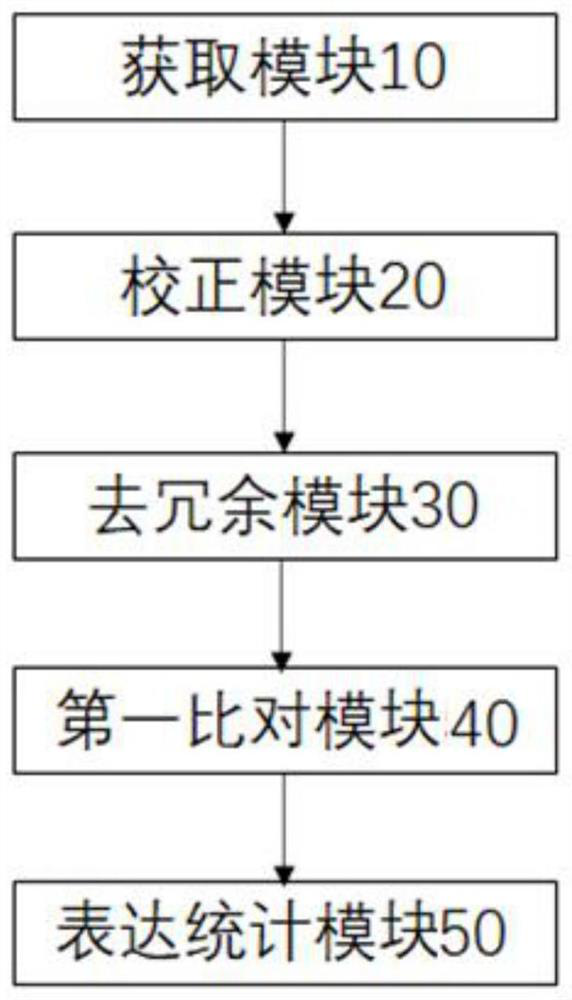
[0086] In this embodiment, a transcriptome analysis system without a reference genome sequence is provided, such as image 3 As shown, the analysis system includes: an acquisition module 10, a correction module 20, a de-redundancy module 30, a first comparison module 40 and an expression statistics module 50, wherein,
[0087] An acquisition module 10, configured to acquire effective sequencing data of the second-generation RNA and effective sequencing data of the third-generation RNA of the sample to be tested;
[0088] The correction module 20 is used to correct the effective sequencing data of the third generation RNA by using the effective sequencing data of the second generation RNA, and obtain the effective data of the third generation correction;
[0089] The de-redundancy module 30 is used to de-redundantly obtain the unigene sequence for the three generations of corrected valid data;
[0090] The first comparison module 40 is used to perform sequence comparison on th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com