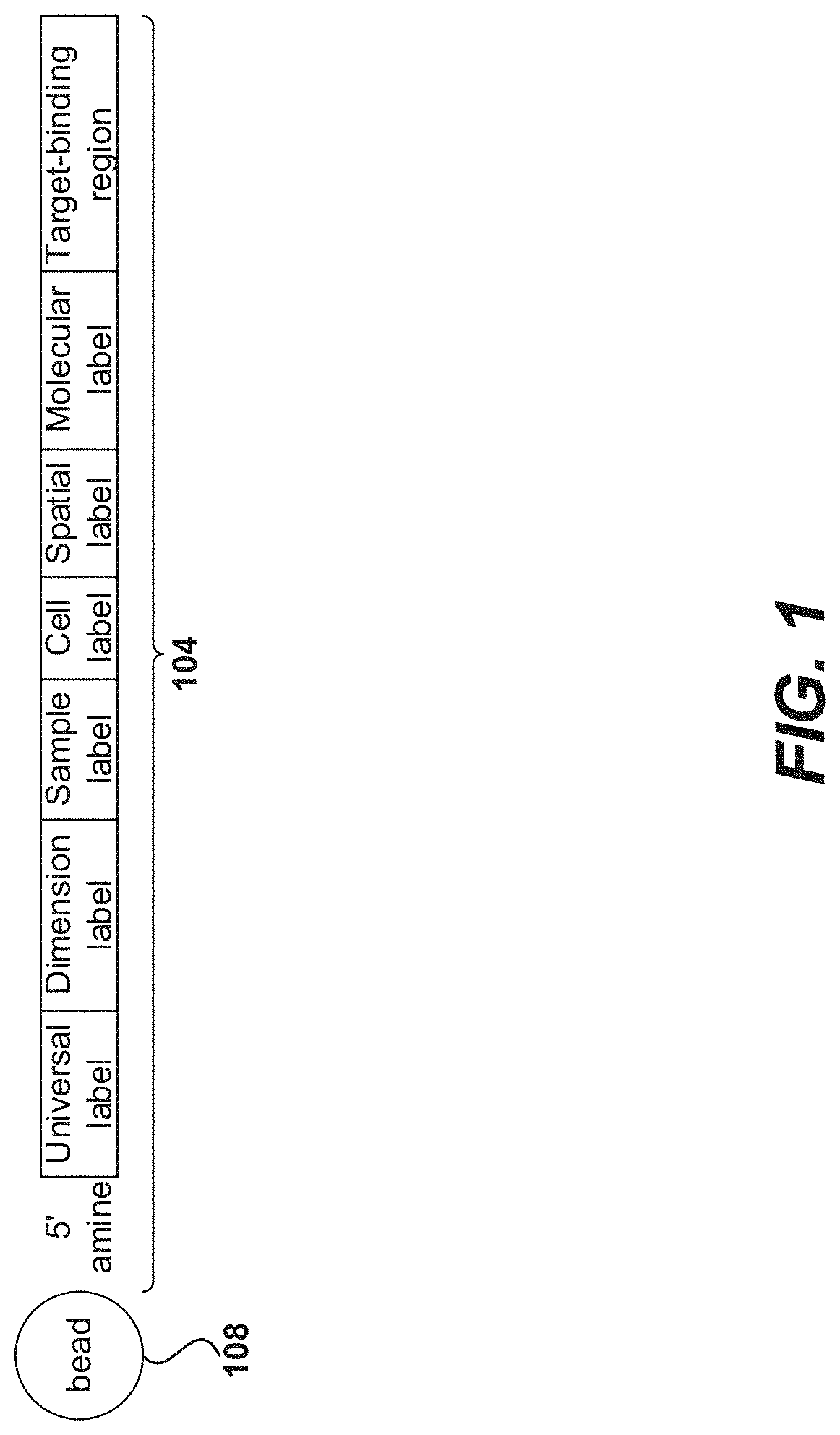
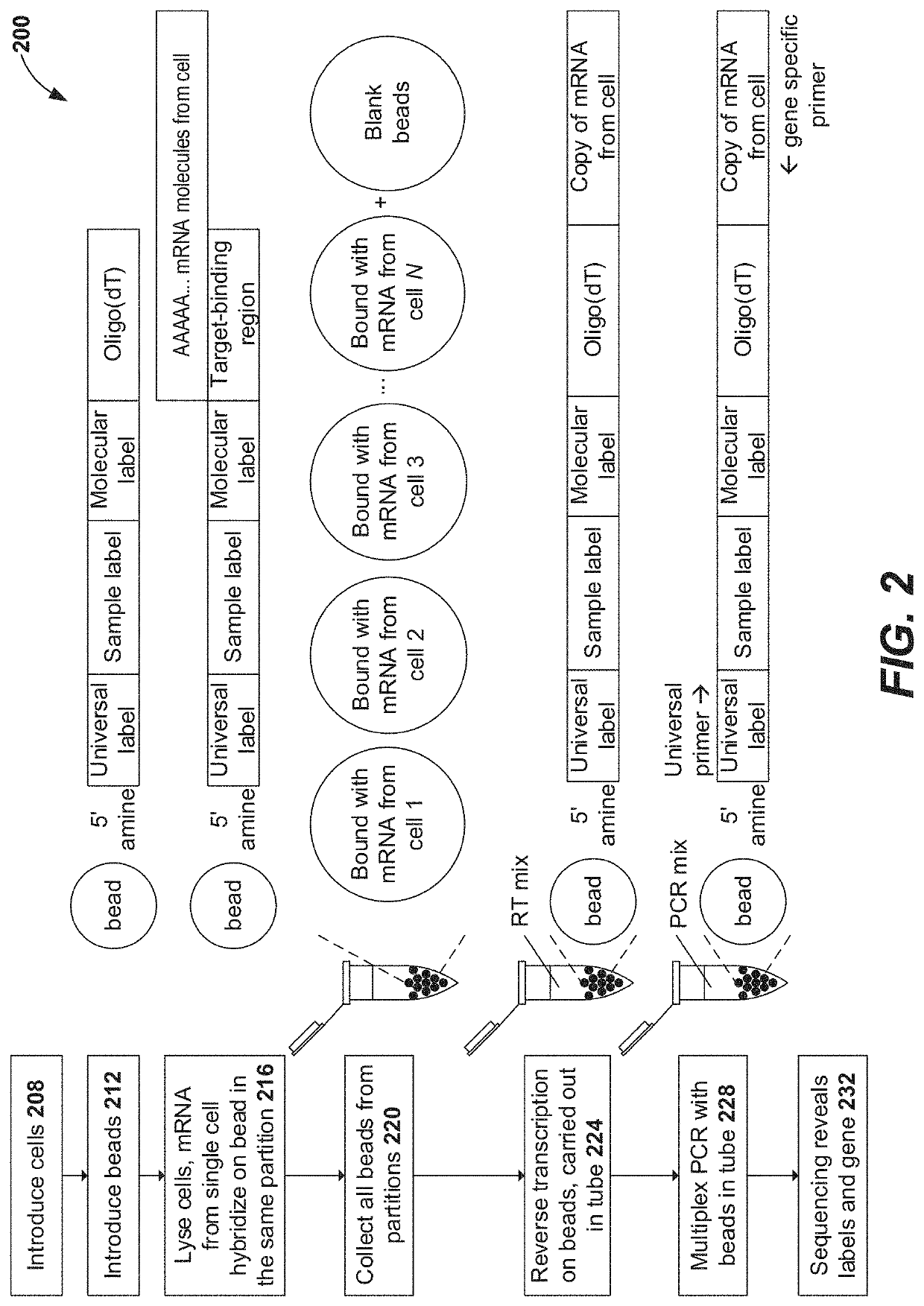
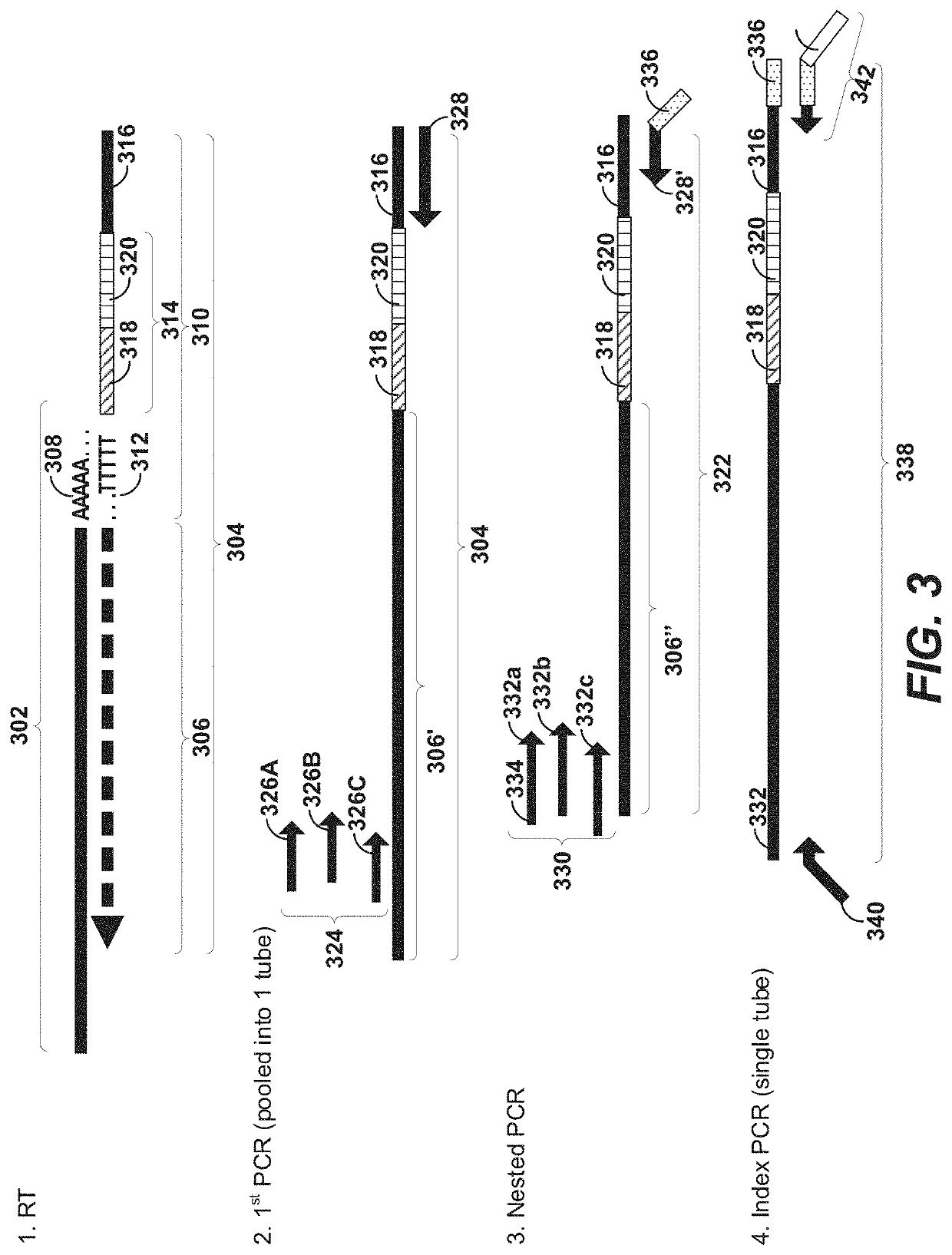
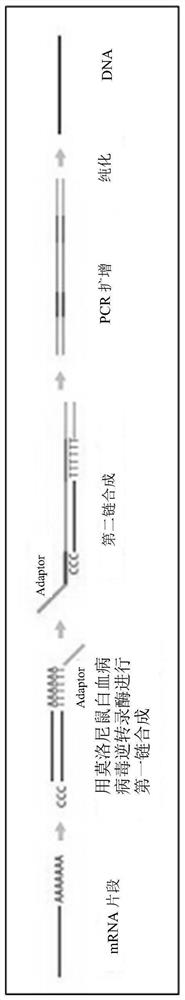
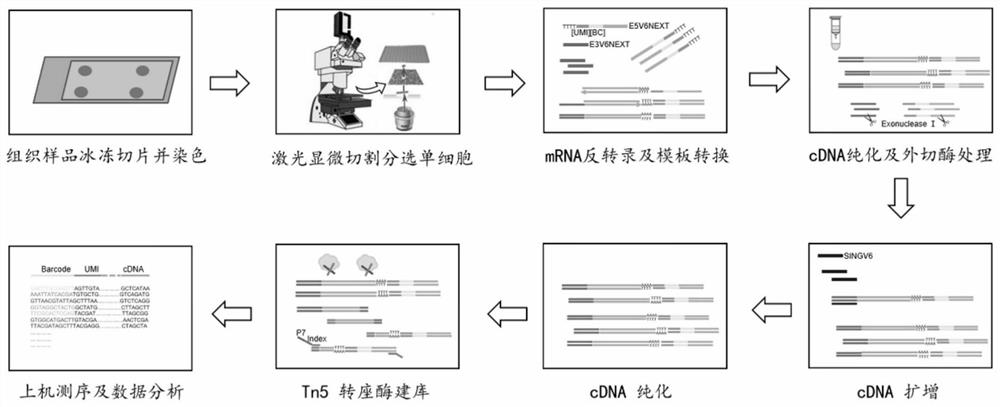
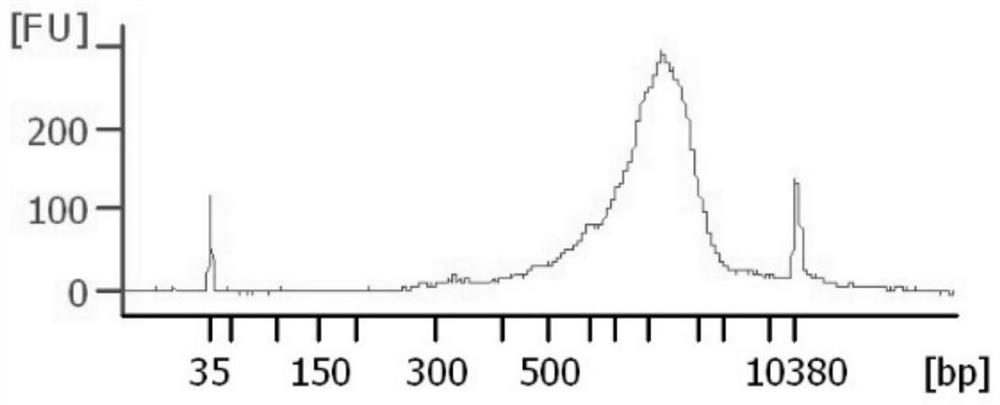
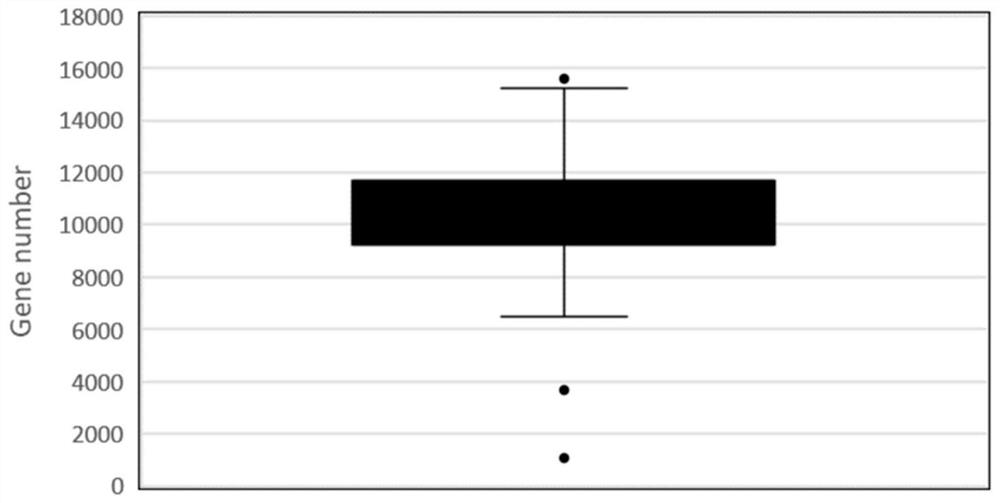
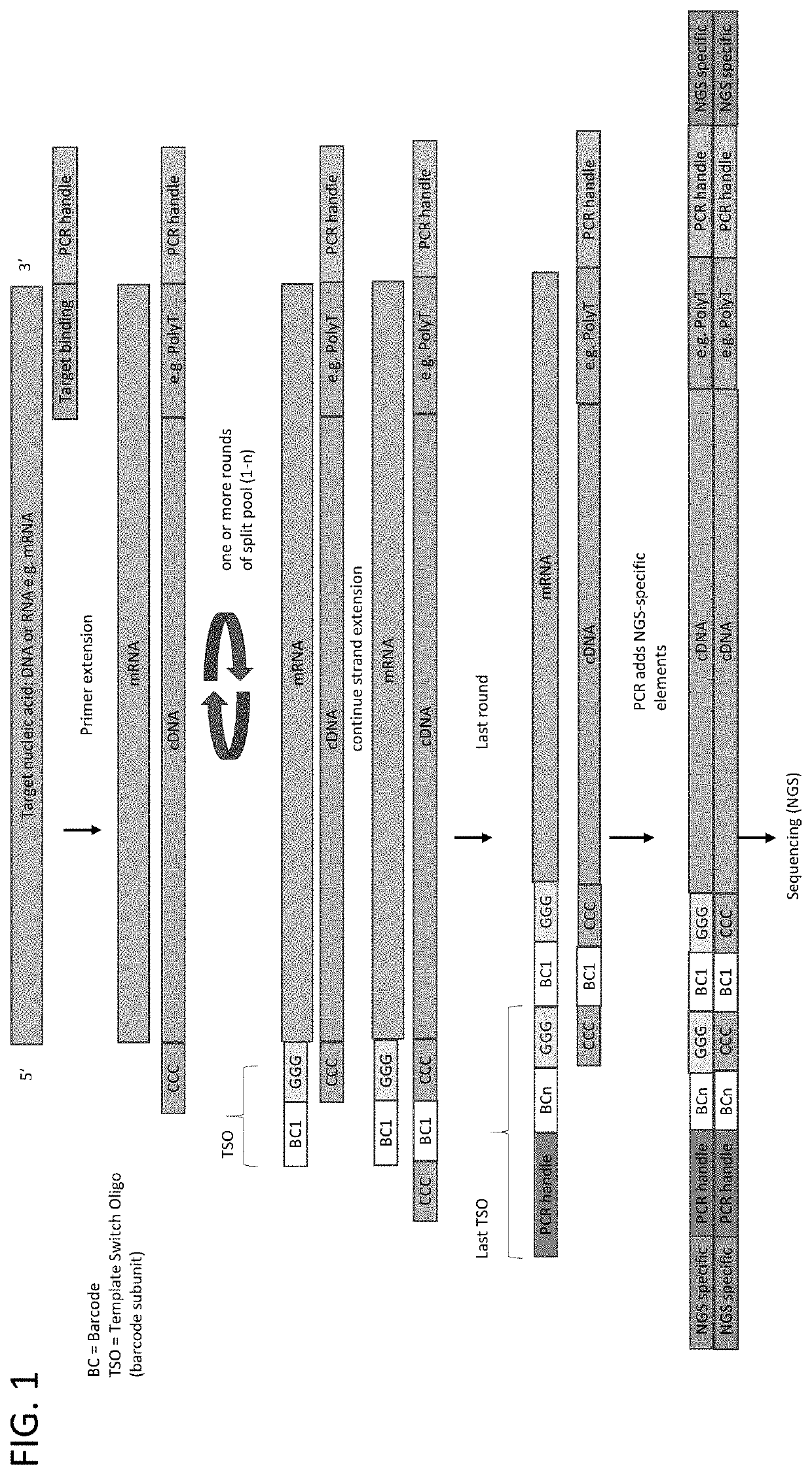
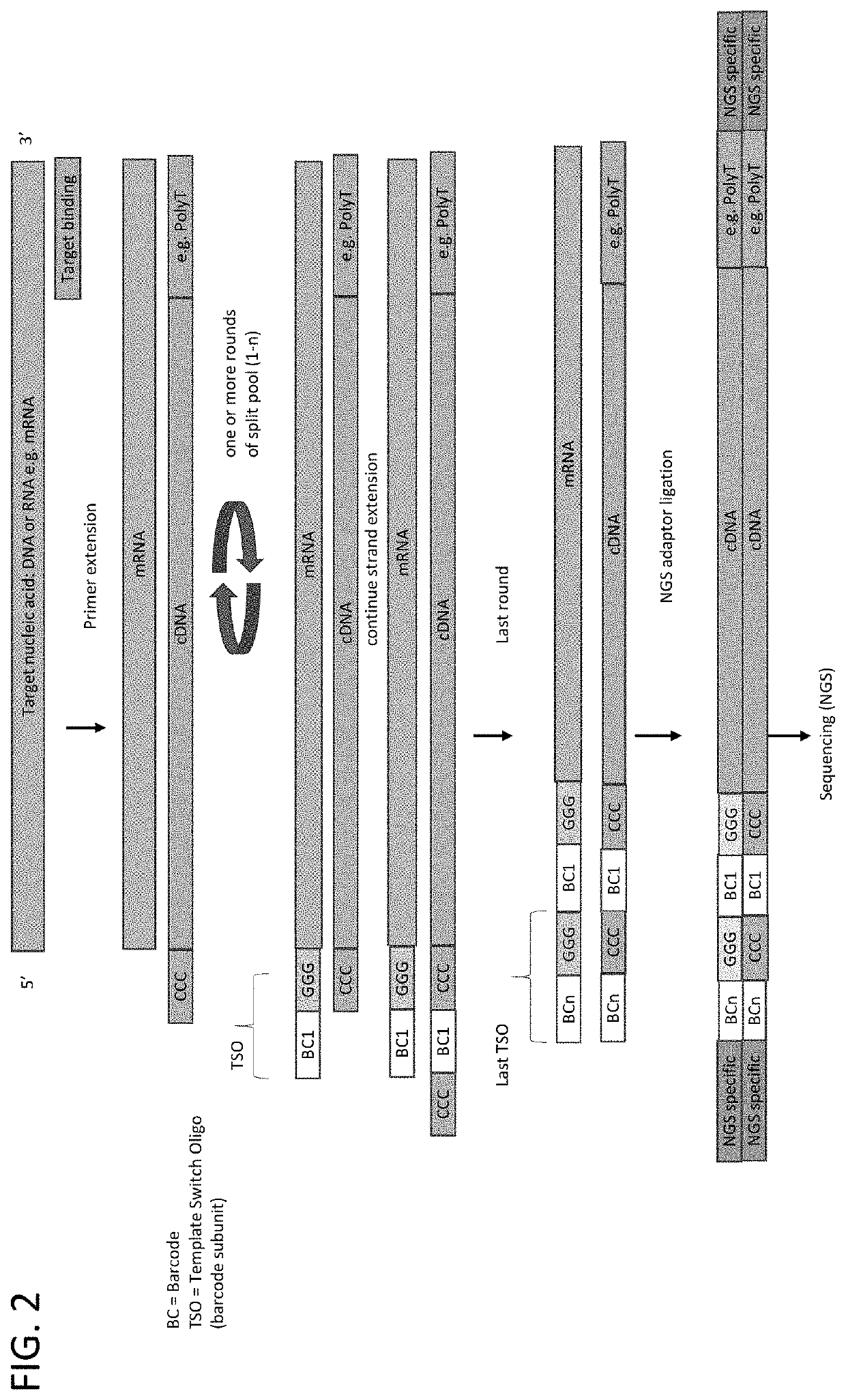
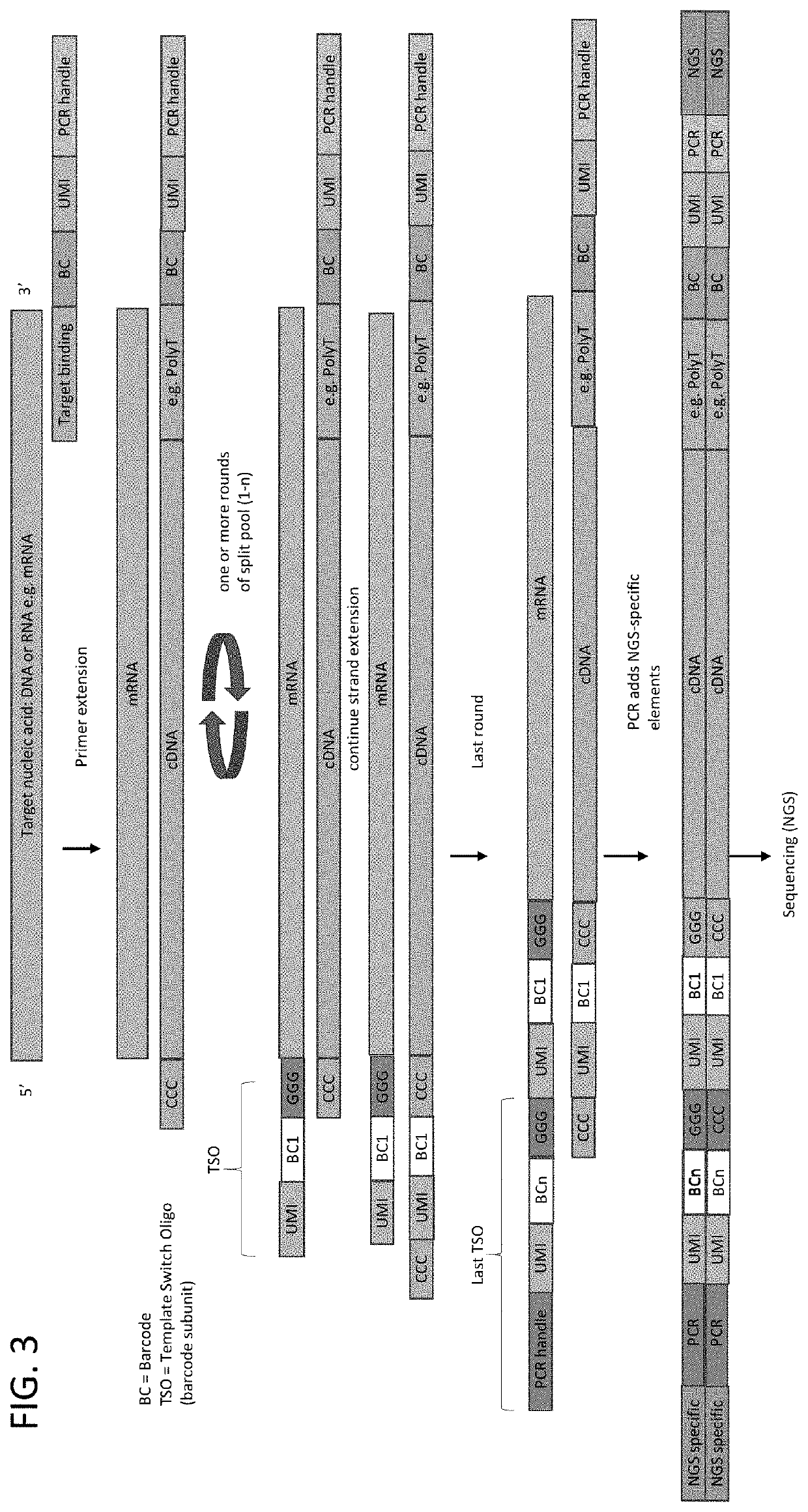
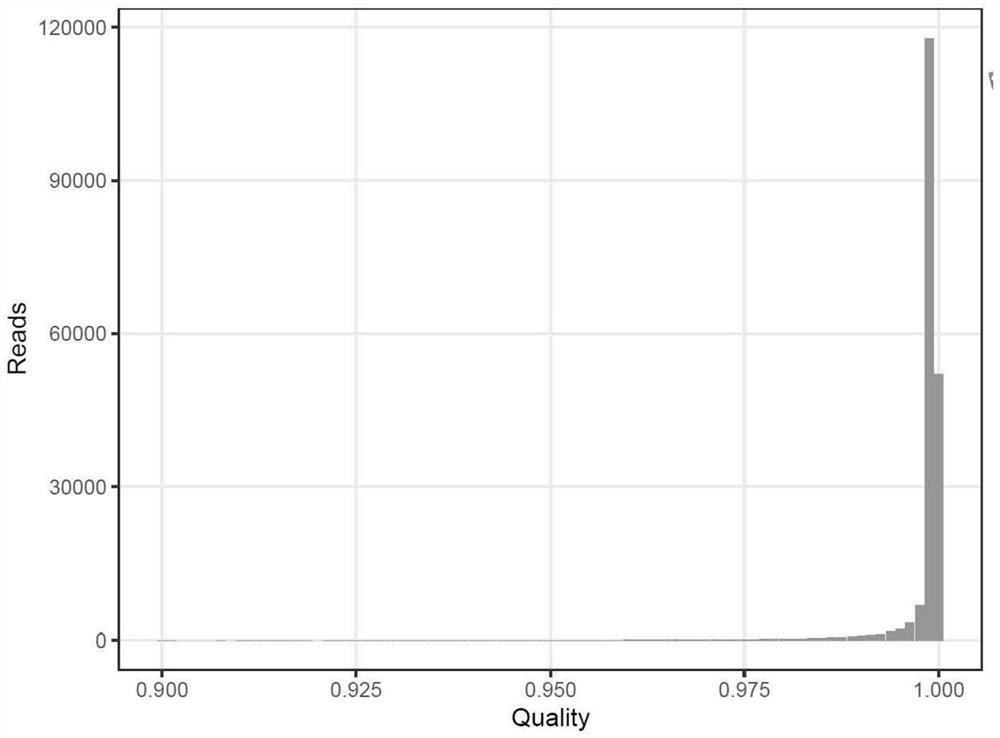
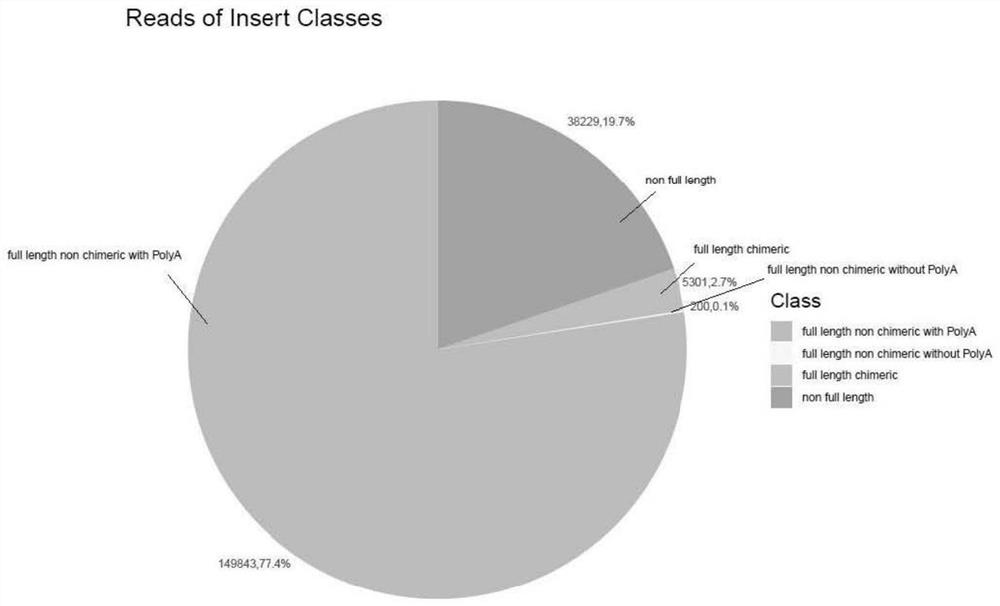
Patents
Literature
53 results about "Transcriptome profiling" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
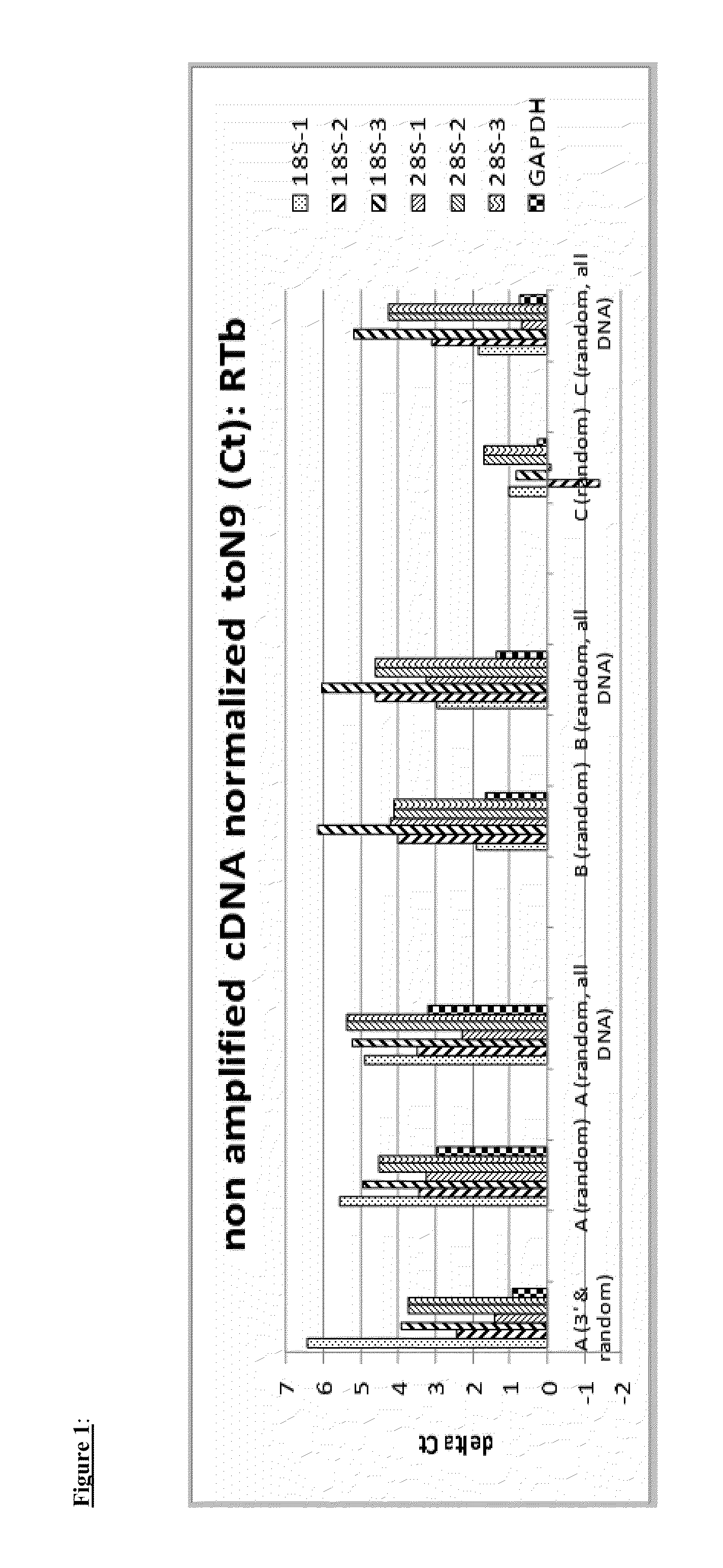
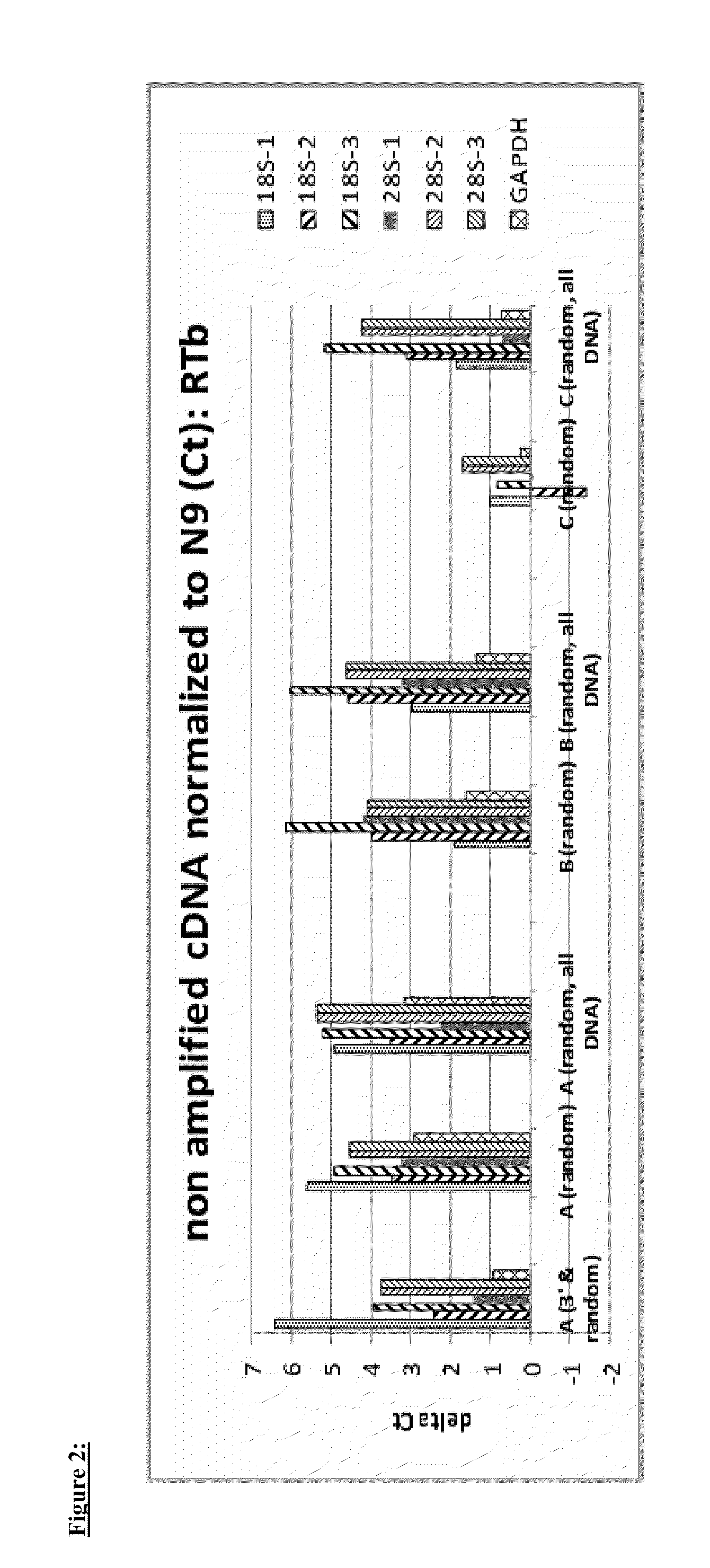
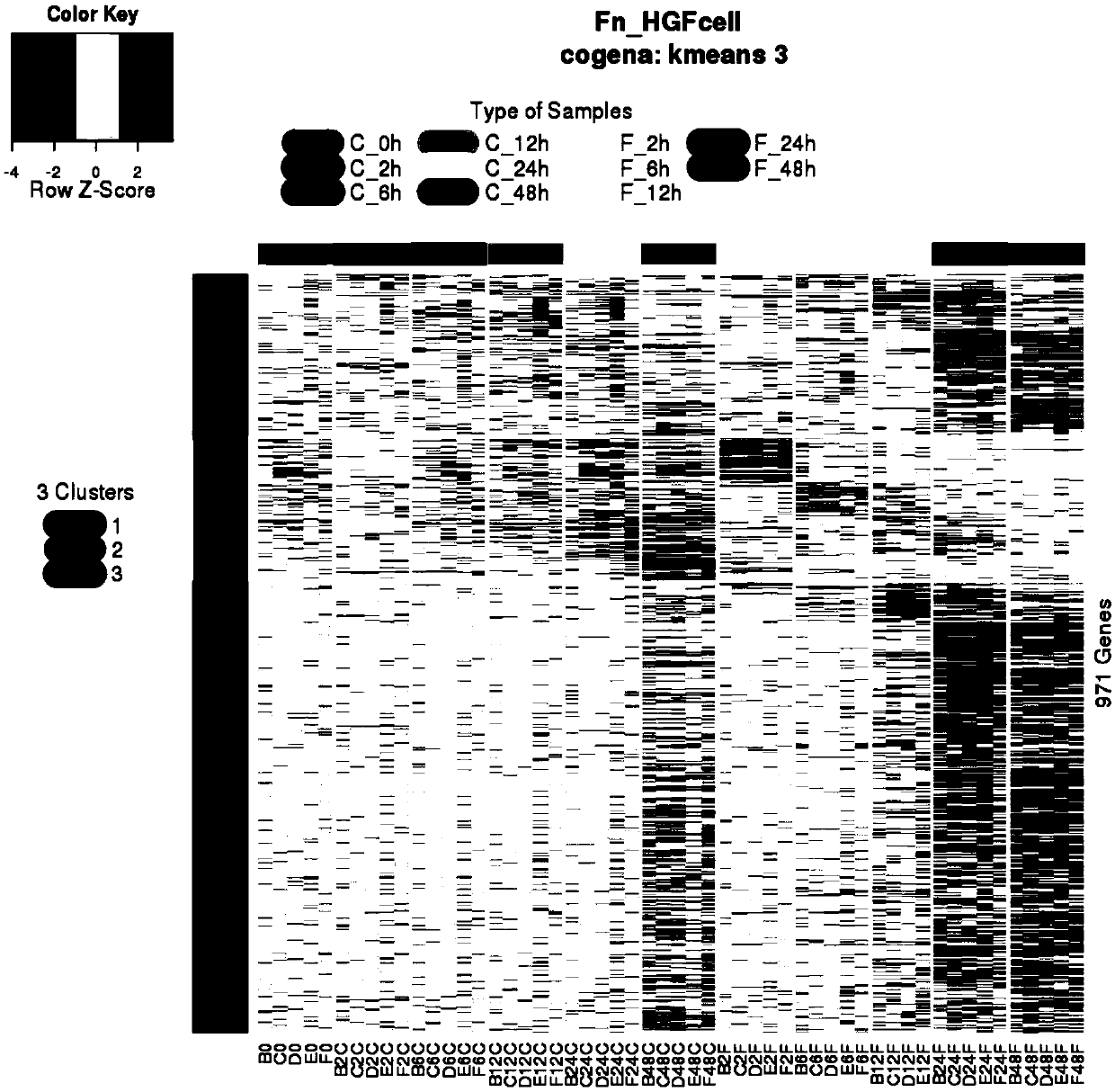
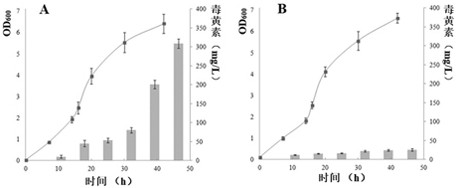
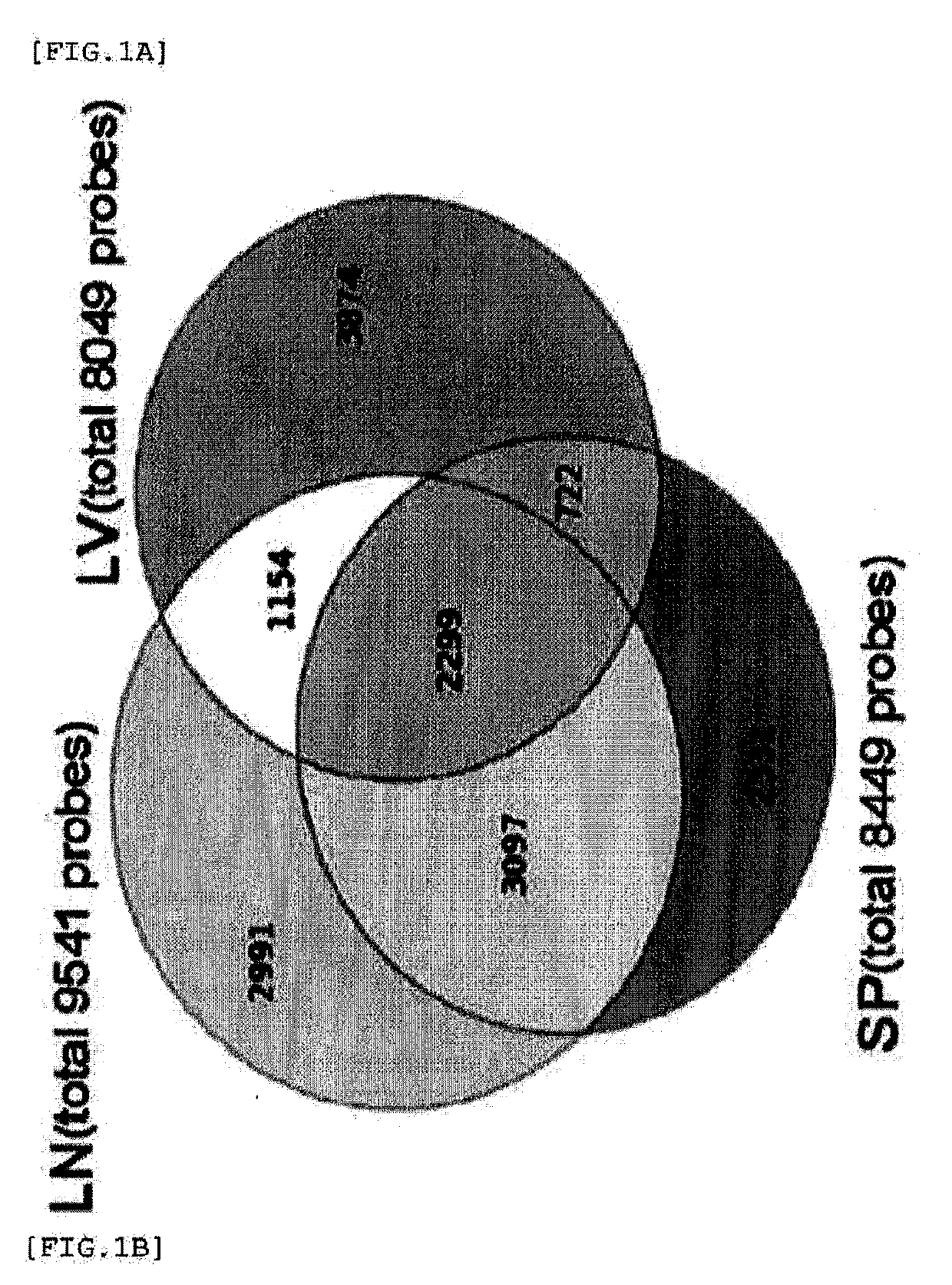
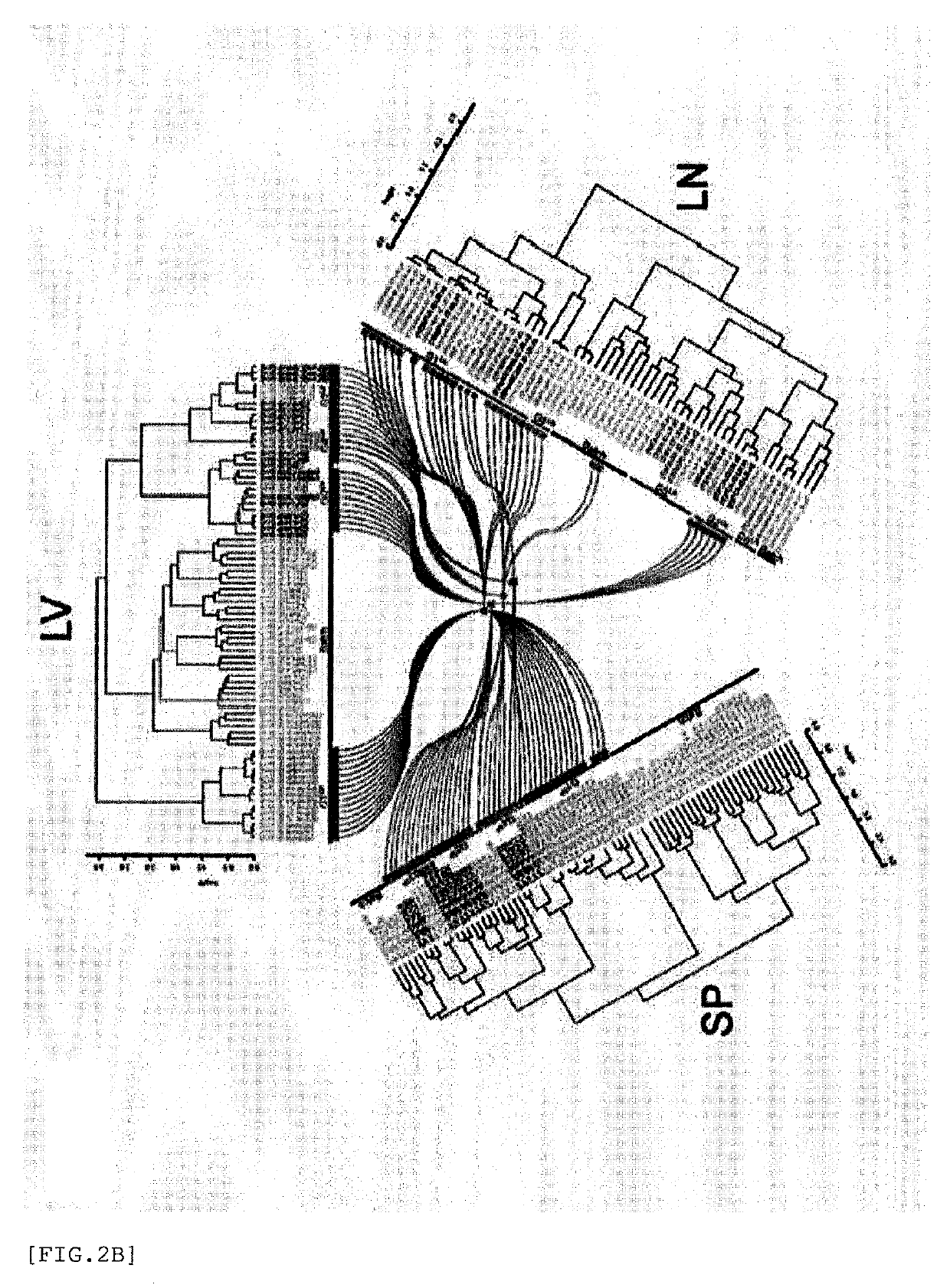
Global transcriptional profiling is a powerful tool that can expose expression patterns to define cellular states or to identify genes with similar expression patterns. In recent years, transcriptome profiling has been widely used to understand the genetic regulation of a particular cell type.
Compositions and methods for whole transcriptome analysis
InactiveUS20110189679A1Reduction and substantial eliminationMicrobiological testing/measurementFermentationRibosomal RNABioinformatics
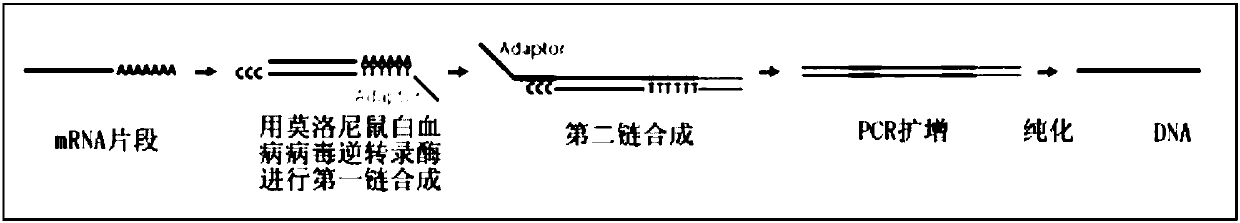
The present invention provides methods and compositions, including kits, for the generation of cDNA from mRNA with reduced ribosomal RNA representation.
Owner:NUGEN TECH
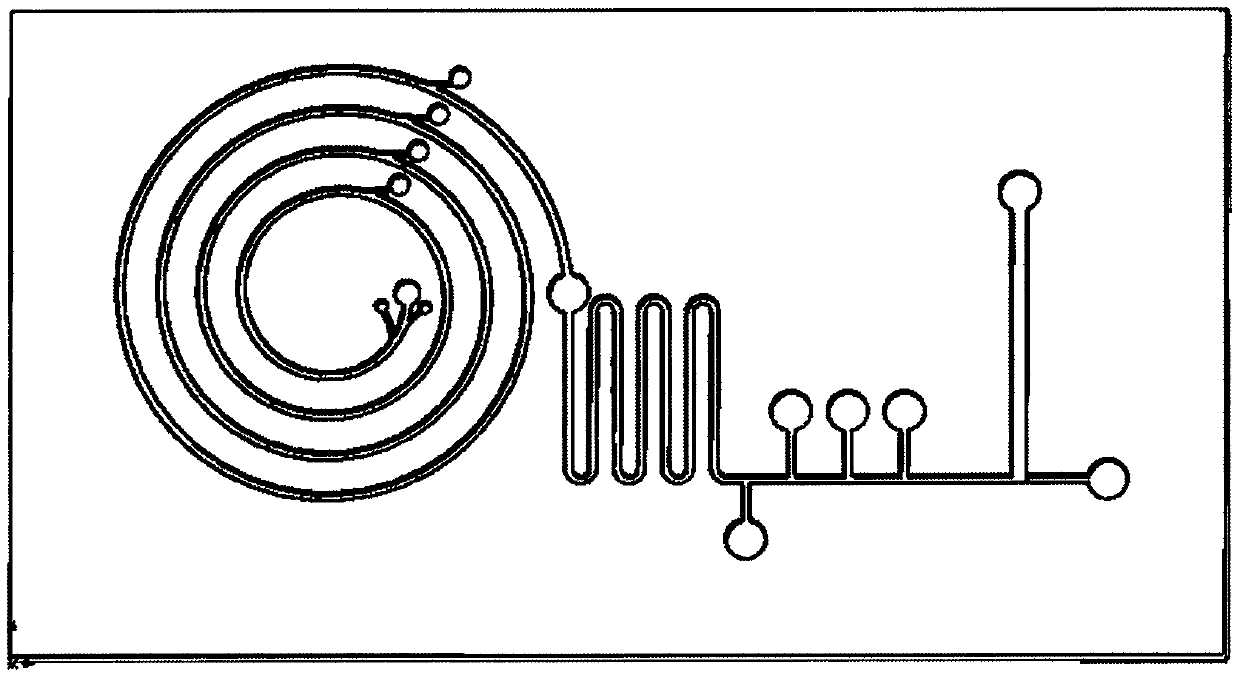
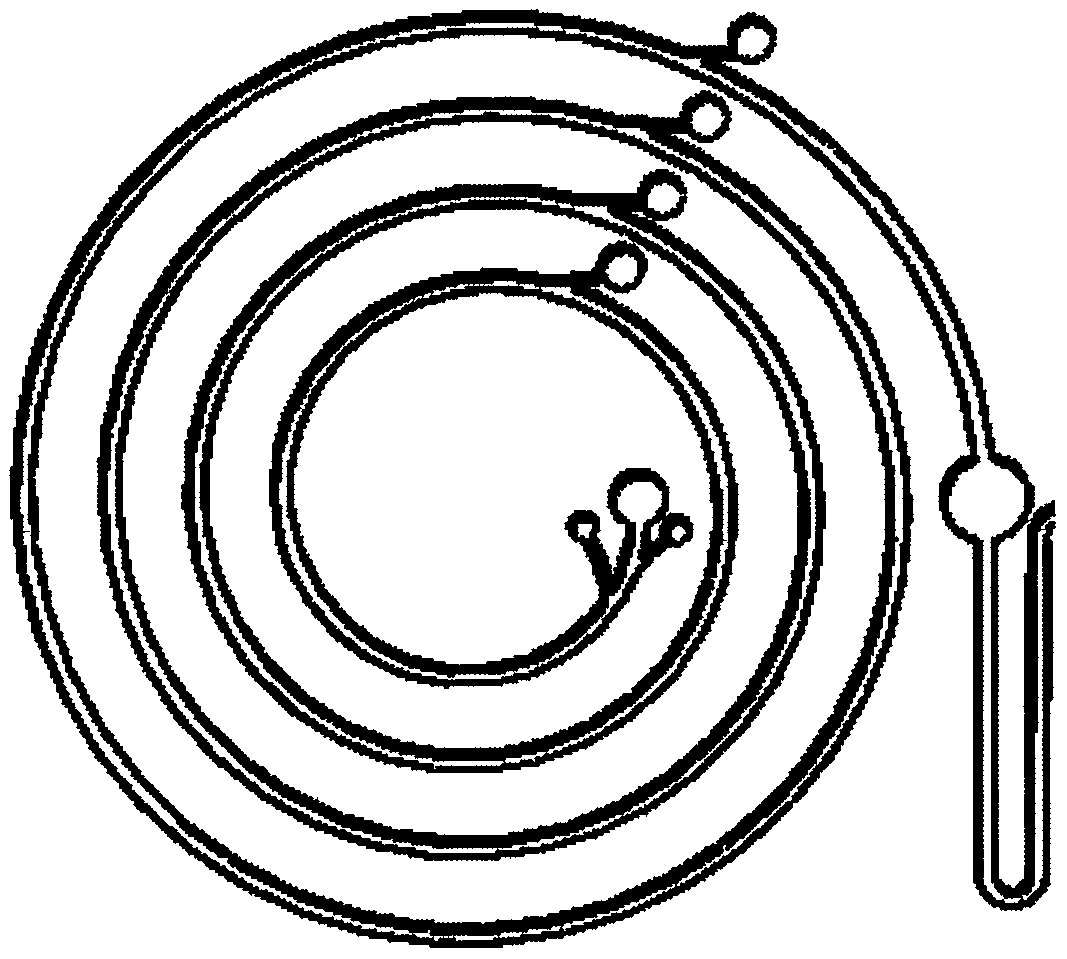
Micro-fluidic chip device for circulating tumor cell (CTC) detection
PendingCN107699478AEasy to operateRealize automated detectionBioreactor/fermenter combinationsBiological substance pretreatmentsStainingWhite blood cell
The invention provides a micro-fluidic chip device for circulating tumor cell (CTC) detection. According to the device, through an integrated micro-fluidic chip mode, functions of blood sample pretreatment, red blood cell lysis, leukocyte screening, circulating tumor cell enrichment, cell marker staining, cleaning and detection, CTC collection and the like are realized. Various functional units ofthe micro-fluidic chip are corresponding to the chip structure. Each solution tanks are connected through microfluidic channels, diameter of the channel matches the structure of researched cells, andfunctional transport of a cell solution in the channel is guaranteed. The device of the invention is used for screening, detecting and collecting circulating tumor cells in peripheral blood sample solutions of patients, providing clinic treatment and guidance and conducting quantitative statistics of circulating tumor cells, and also can be used for downstream single-cell genome and transcriptomeanalysis. The device of the invention is simple to operate, has advantages of saving time cost, realizing automatic detection and raising detection efficiency and analysis efficiency, and has vitallyimportant clinical examination value.
Owner:朱嗣博
Single-cell RNA (ribonucleic acid) reverse transcription and library construction method
ActiveCN108103055AEfficient and uniform transcriptAvoid pollutionMicrobiological testing/measurementLibrary creationCDNA libraryTotal rna
The invention belongs to the field of transcriptome analysis and relates to a quick single-cell RNA (ribonucleic acid) reverse transcription and library construction method. According to the method, 20-500ng high-quality full-length double-chain cDNA (complementary deoxyribonucleic acid) is amplified by taking 1-2000 cells or 10pg-20ng extracted eukaryote total RNA as an initial, and a high-quality cDNA library meeting downstream analysis requirements is obtained. The method effectively avoids 3' preference and genome DNA contamination in a cDNA synthesis process; an expression quantitation molecule label can assist in gene expression quantity calculation; and at the same time, the expression quantitation molecule label can also keep chain source information during complete amplification of RNA sequence information. The method can achieve a reverse transcription and amplification library construction success rate of above 95%; the cDNA library can be connected with an Illumina main stream sequencing platform; lower computer data (5M Reads) can detect gene expression of above 90%; the gene expression consistency exceeds 90%; amplification has no obvious bias; and a required sample input amount is smaller.
Owner:XUKANG MEDICAL SCI & TECH (SUZHOU) CO LTD
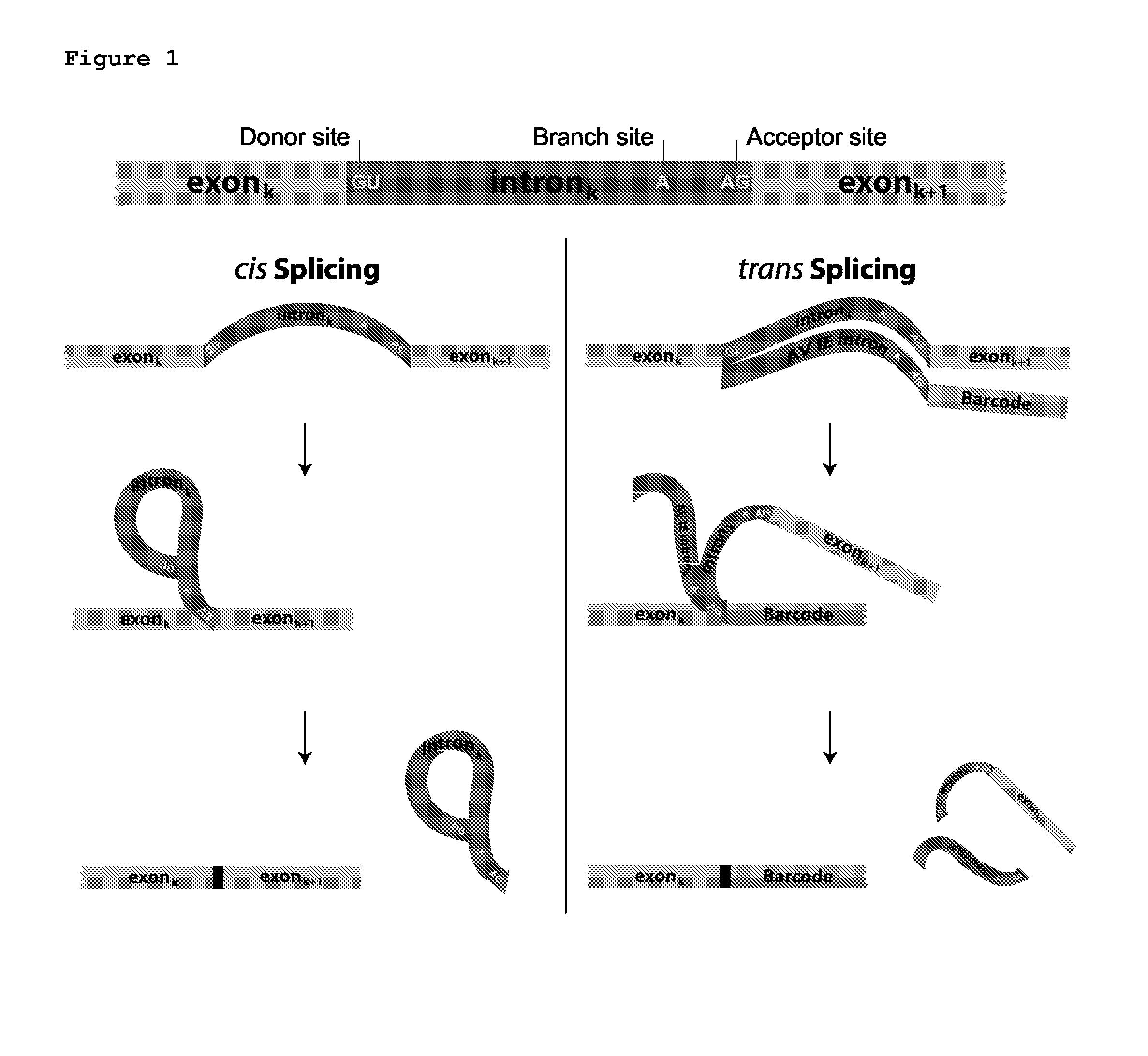
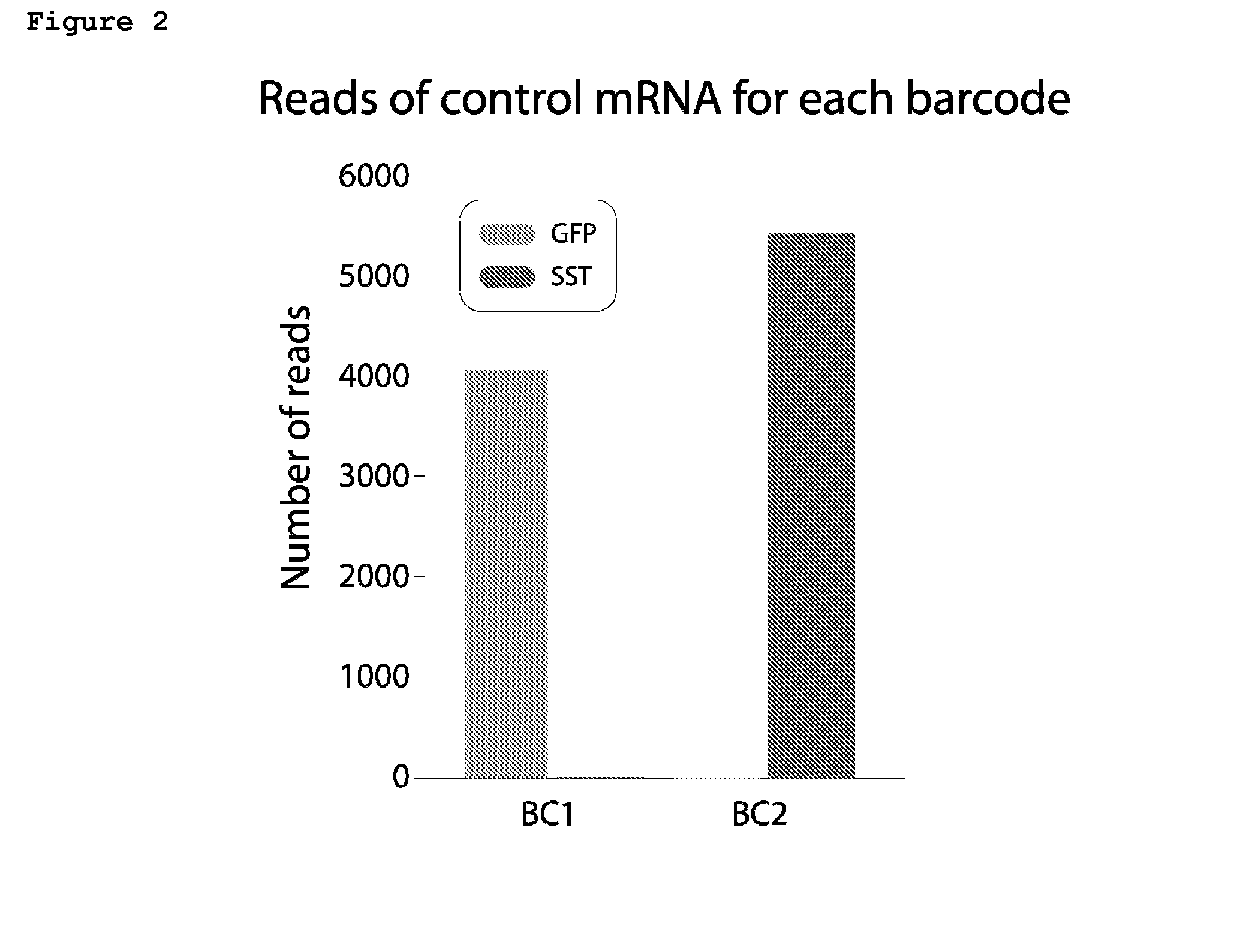
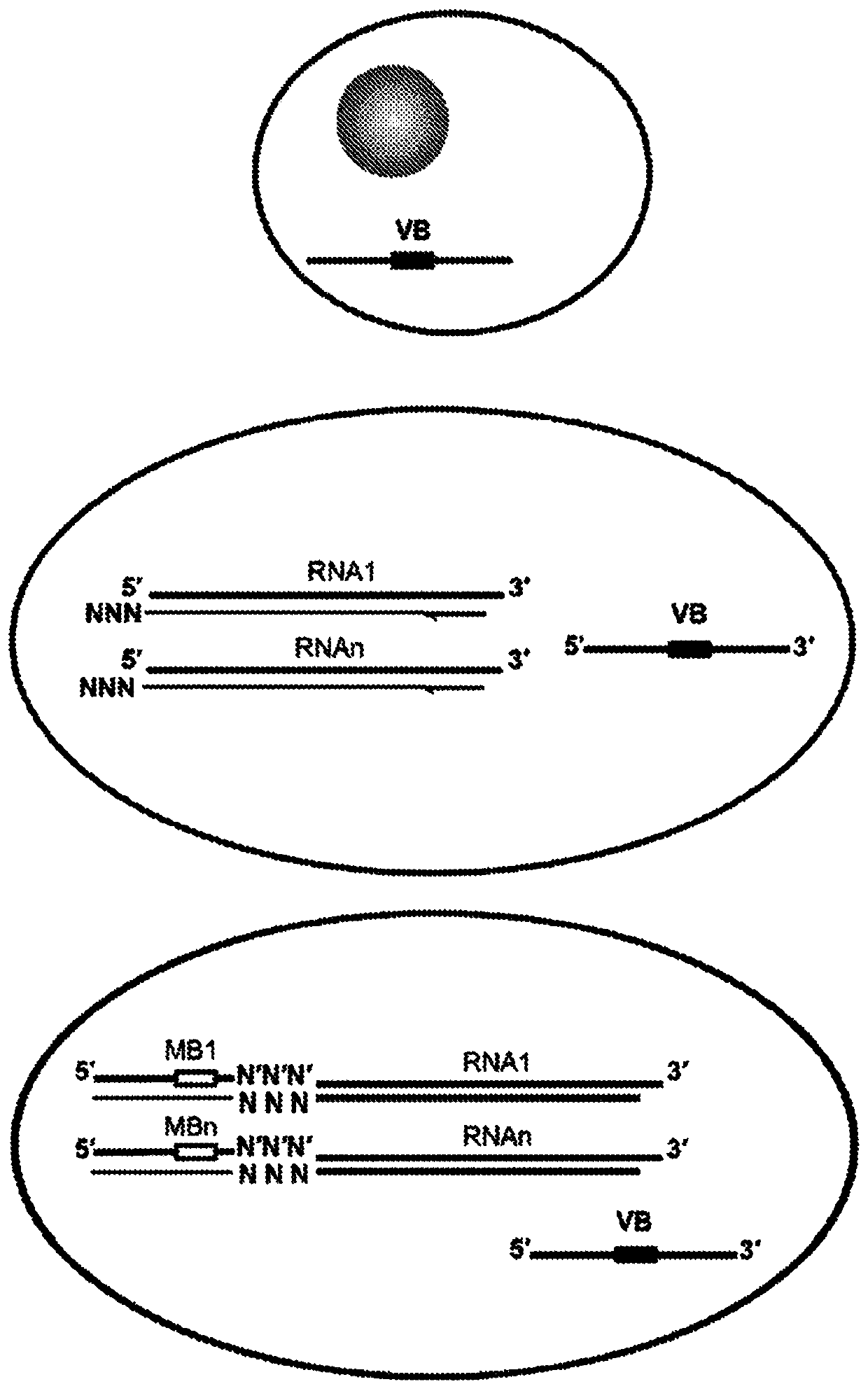
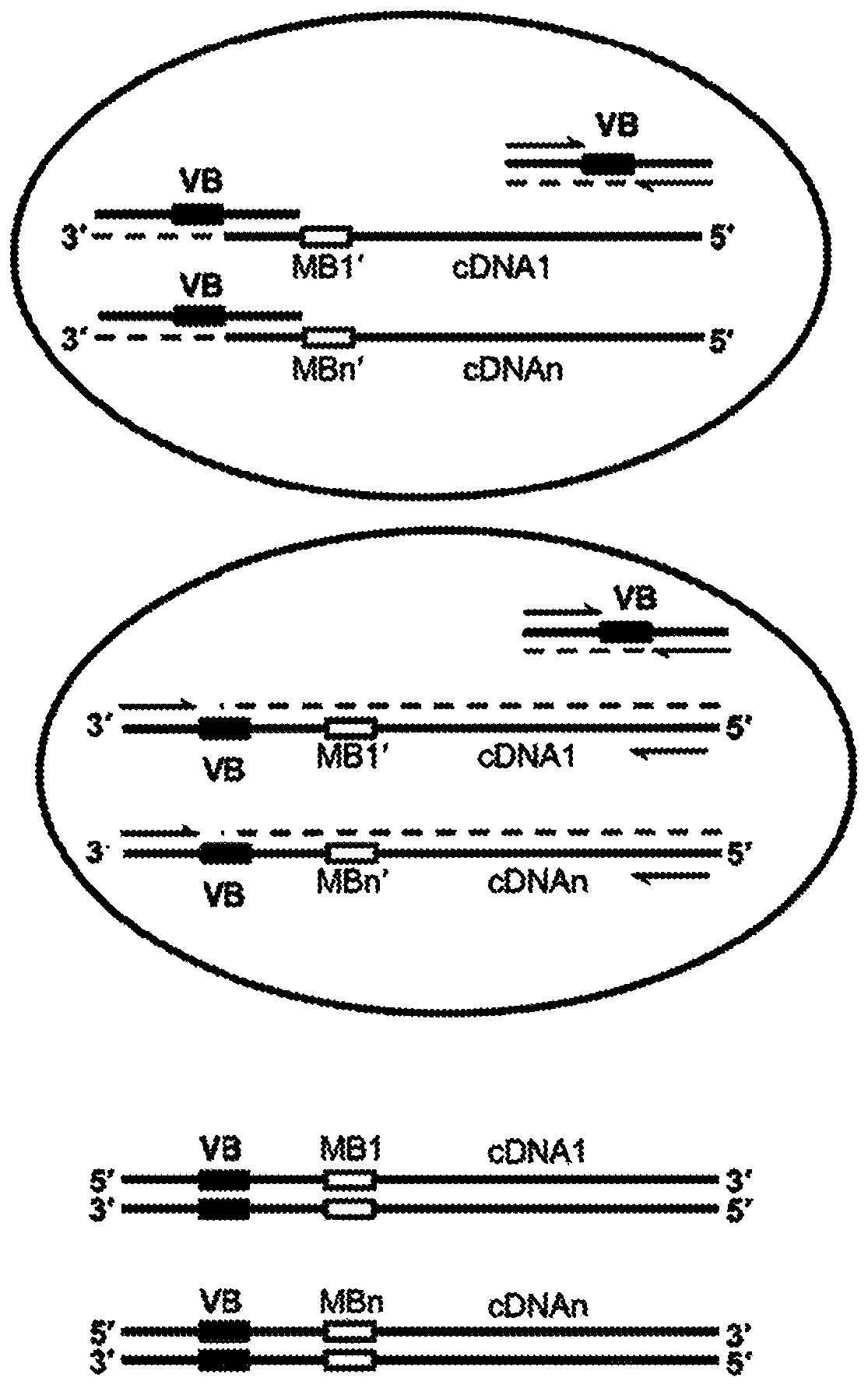
Trans-splicing transcriptome profiling
The present invention provides a method of identifying mRNA transcripts in the transcriptome of a cell comprising i) delivering into the cell a donor expression vector comprising nucleotides in a sequence encoding a trans-splicing barcode cassette, wherein the trans-splicing barcode cassette comprises a) a first portion, the nucleotide sequence of which encodes an intron comprising as part of its 3′ end, or followed at its 3′ end by a splice-site nucleotide sequence; followed at its 3′ end by, b) a second portion, the nucleotide sequence of which encodes a barcode polynucleotide; followed at its 3′ end by c) a third portion, which encodes a nucleotide identification element sequence, ii) exposing the cell to conditions such that the cell produces multiple copies of the trans-splicing barcode cassette encoded by the donor expression vector, which multiple copies of the trans-splicing barcode cassette each splice the barcode polynucleotide onto a mRNA transcript of the cell, thereby forming multiple mRNA transcripts of the cell, each spliced to the barcode polynucleotide; and iii) identifying the multiple mRNA transcripts that are spliced to the barcode polynucleotides, thereby identifying mRNA transcripts in the transcriptome of the cell.
Owner:COLD SPRING HARBOR LAB INC
Methods and Compositions for Activity Dependent Transcriptome Profiling
InactiveUS20150141274A1Bioreactor/fermenter combinationsBiological substance pretreatmentsCell typeMolecular biology
Owner:THE ROCKEFELLER UNIV
Mesophilic DNA polymerase extension blockers
PendingUS20210238661A1Reduce generationMicrobiological testing/measurementGenomic cloneOligonucleotide
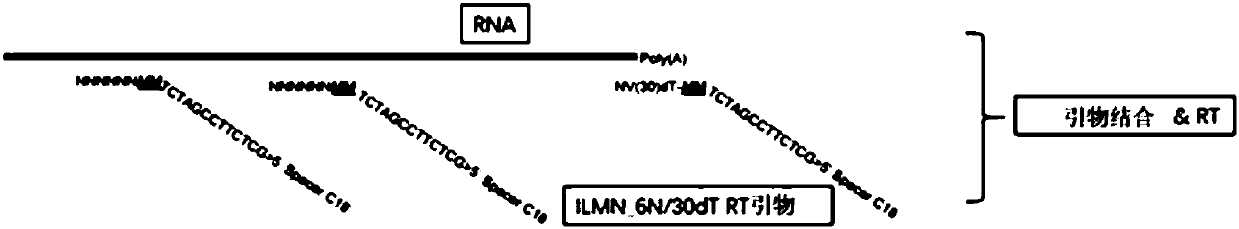
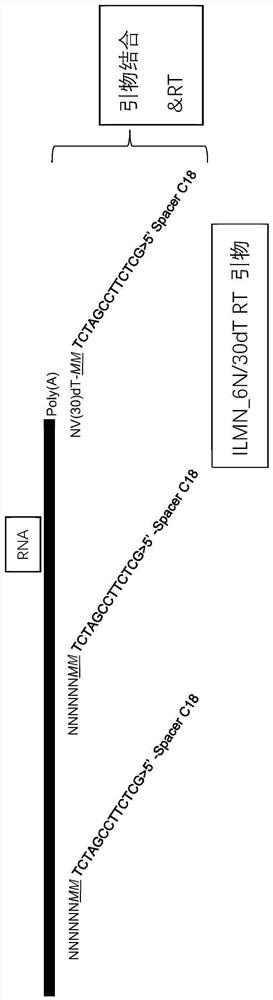
Disclosed herein include systems, methods, compositions, and kits for 5′-based gene expression profiling and for whole transcriptome analysis (WTA) with random priming and extension (RPE). Blocker oligonucleotides capable of specifically binding to a portion of an oligonucleotide barcode are provided in some embodiments. The blocker oligonucleotides can reduce of the generation of undesirable extension products, such as, for example, the extension products of random primers hybridized to a portion of the oligonucleotide barcode. Immune repertoire profiling methods are also provided in some embodiments.
Owner:BECTON DICKINSON & CO
Application of piperlongumine in the preparation of anti-bacterial and anti-inflammatory drugs
InactiveCN109602744AHas antibacterial and anti-inflammatory effectsBroaden the use of pharmaceuticalsAntibacterial agentsOrganic active ingredientsAnti-inflammatoryAnti bacterial
The invention discloses an application of piperlongumine in the preparation of anti-bacterial and anti-inflammatory drugs. By performing RNA-seq transcriptome analysis for the cells of people infectedwith bacteria, a list of drug candidates with functions of antisepsis and anti-inflammation can be obtained on account of analysis methods of co-expression pathway and drug gene aggregation, whereinthe list comprises a plurality of marketed drugs with anti-bacterial and anti-inflammatory effects, the piperlongumine not discovered to have anti-bacterial and anti-inflammatory effects is right in the list. Therefore, the piperlongumine can be used in the preparation of anti-bacterial and anti-inflammatory drugs.
Owner:GENERAL HOSPITAL OF PLA +1
Methods and compositions for cDNA synthesis and single-cell transcriptome profiling using template switching reaction
InactiveCN105579587ASugar derivativesMicrobiological testing/measurementCDNA librarySingle cell transcriptome
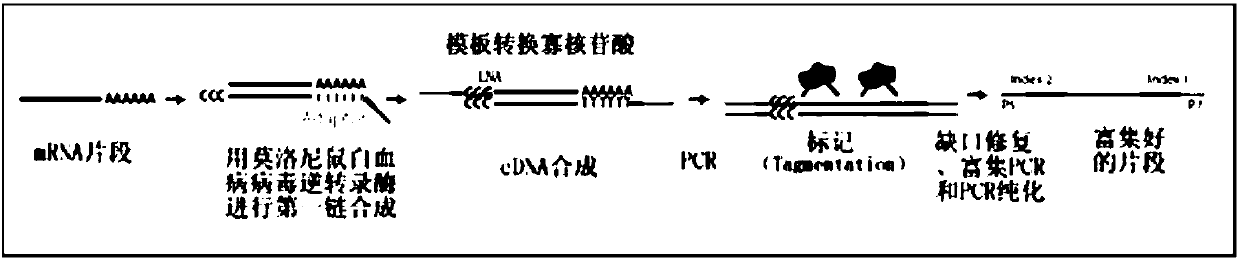
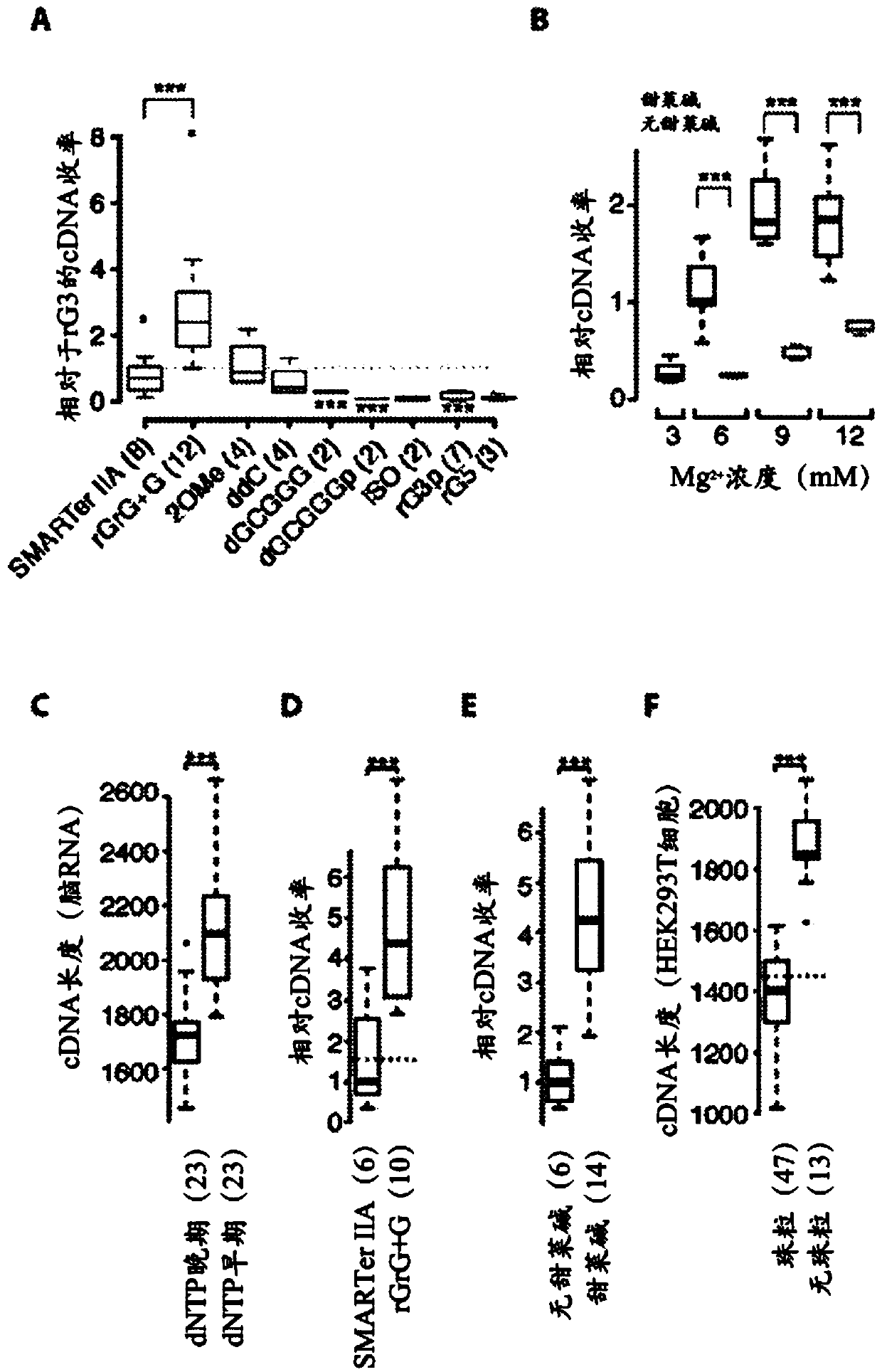
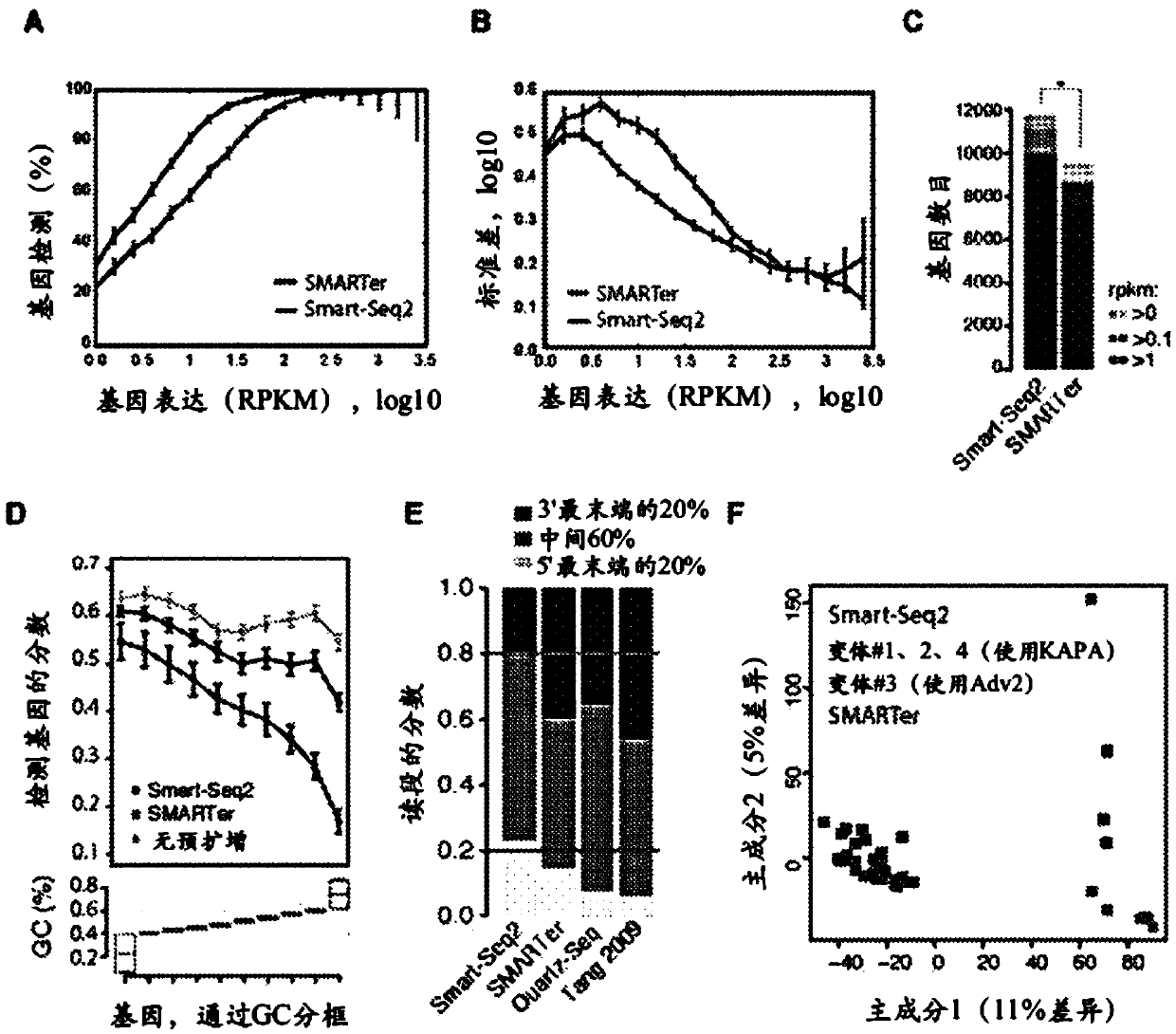
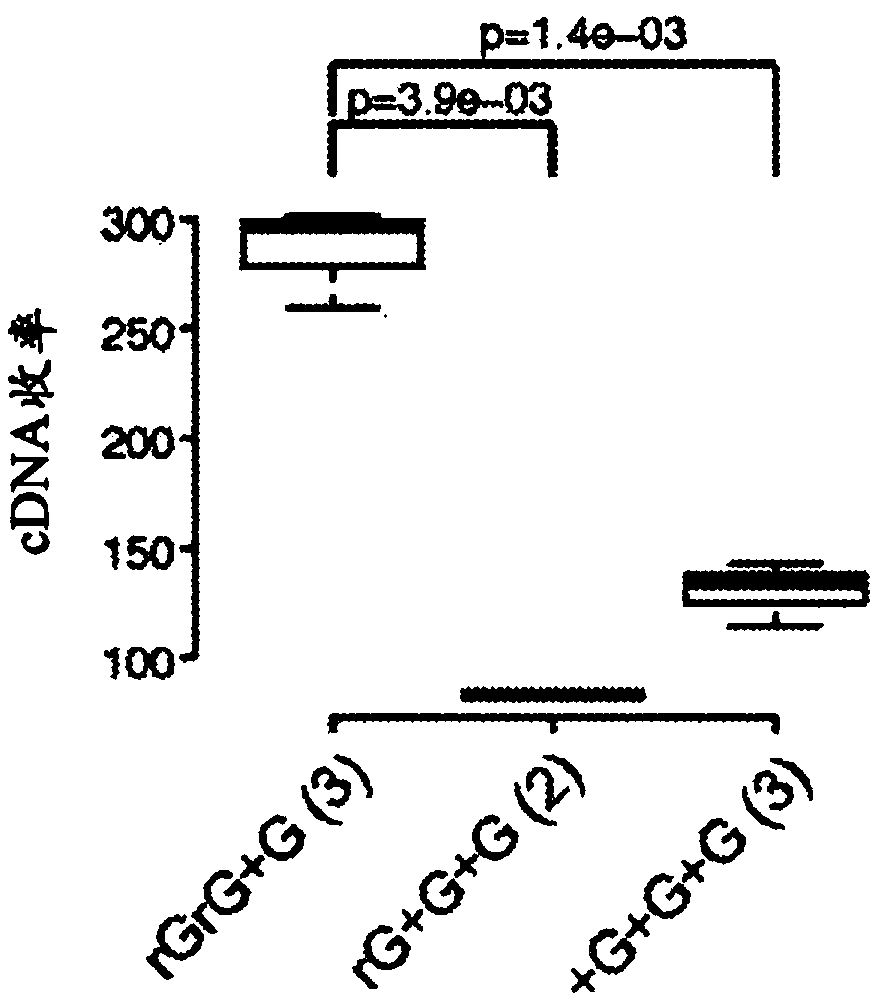
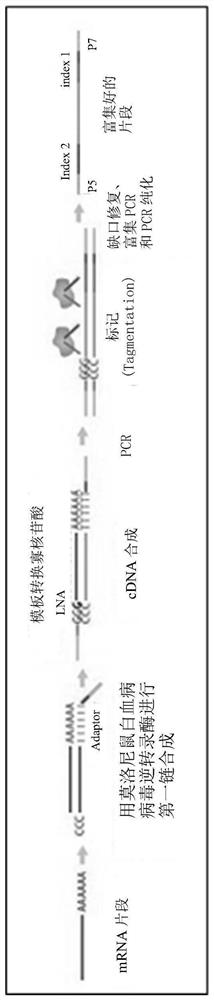
This application discloses methods for cDN'A synthesis with improved reverse transcription, template switching and preamplification to increase both yield and average length of cDNA libraries generated from individual cells. The new methods include exchanging a single nucleoside residue for a locked nucleic acid (INA) at the TSO 3' end, using a methyl group donor, and / or a MgCb concentration higher than conventionally used. Single-cell transcriptome analyses incorporating these differences have full-length coverage, improved sensitivity and accuracy, have less bias and are more amendable to cost-effective automation. The invention also provides cDNA molecules comprising a locked nucleic acid at the 3'-end, compositions and cDNA libraries comprising these cDNA molecules, and methods for single-cell transcriptome profiling.
Owner:LUDWIG INST FOR CANCER RES LTD
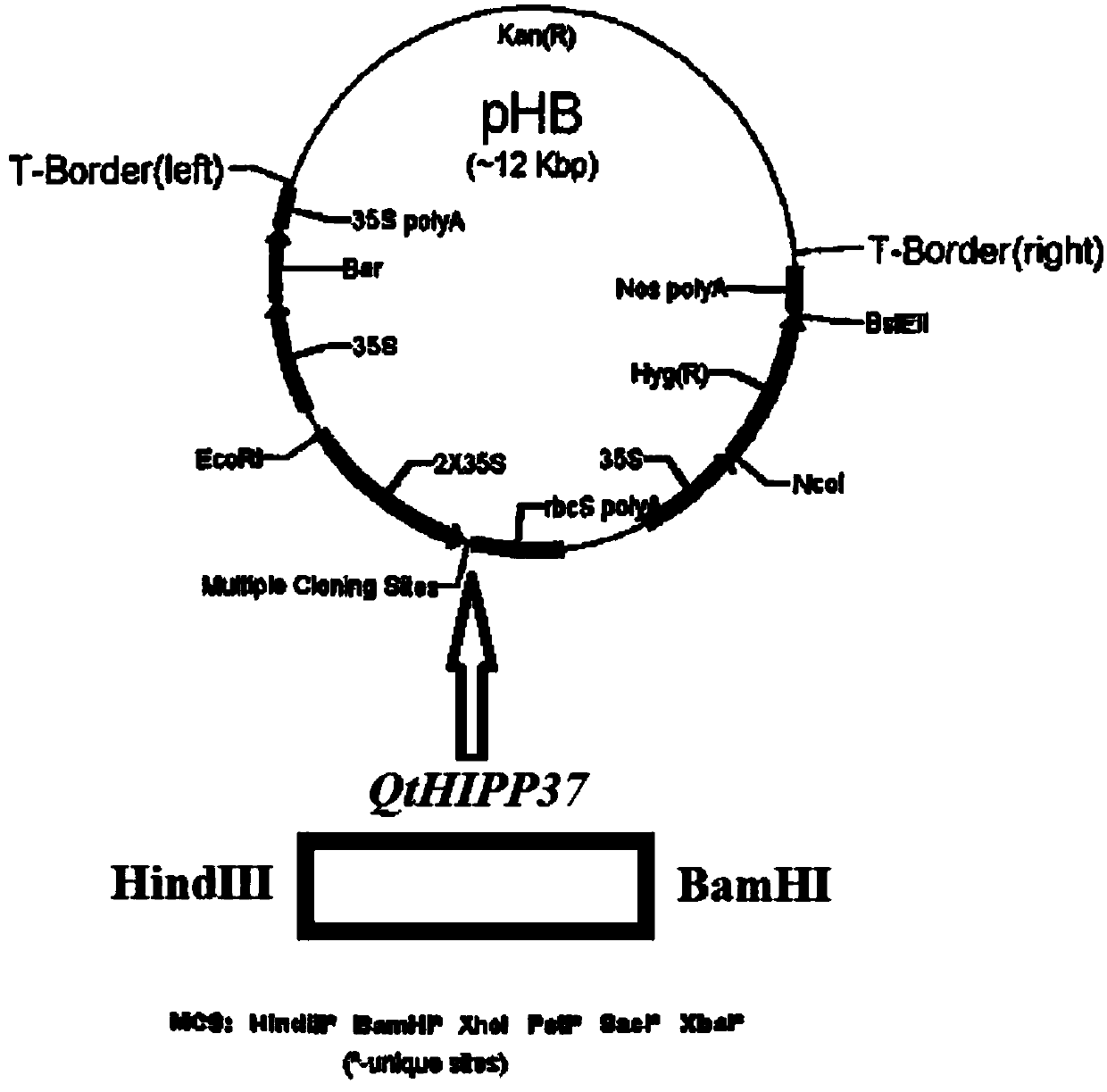
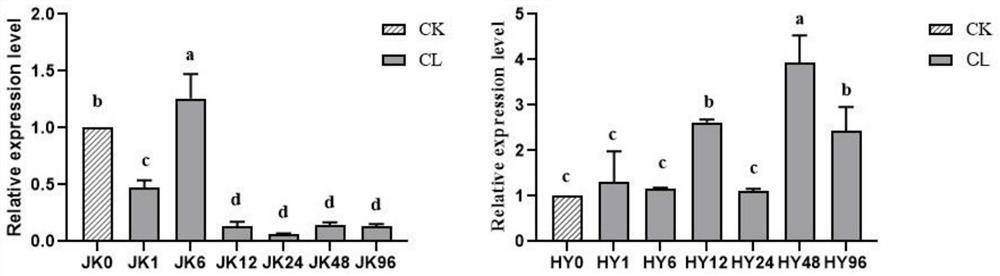
Ornithogalum thyrsoides QtHIPP37 gene and application thereof
ActiveCN109575111ANormal horticultural traitsNormal growth and developmentPlant peptidesFermentationNicotiana tabacumAgricultural science
Owner:四川天艺优境环境科技有限公司 +1
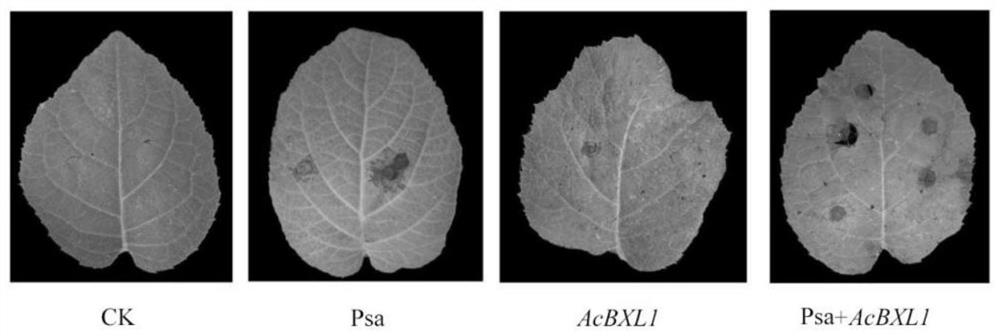
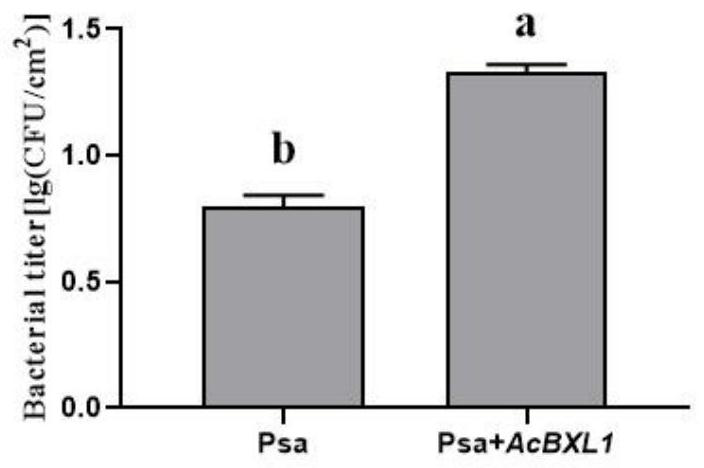
Kiwi fruit canker susceptible gene AcBXL1 and application thereof
ActiveCN113604490ASusceptibleThe promotion effect is obviousFermentationPlant tissue cultureBiotechnologyTransforming Genes
The invention discloses a kiwi fruit canker susceptible gene AcBXL1 and application thereof. The gene AcBXL1 and the sequence of the encoding protein thereof are respectively shown as SEQ ID NO: 1 and 2. According to the invention, through analysis of kiwi fruit genome and transcriptome, the gene sequence of kiwi fruit AcBXL1 is successfully obtained, and through transient expression and stable transformation genetic transformation experiments, the gene AcBXL1 is proved to have the function of promoting the susceptibility of kiwi fruit to canker, thus laying the foundation for creating new disease-resistant varieties for follow-up gene editing and controlling canker.
Owner:ANHUI AGRICULTURAL UNIVERSITY
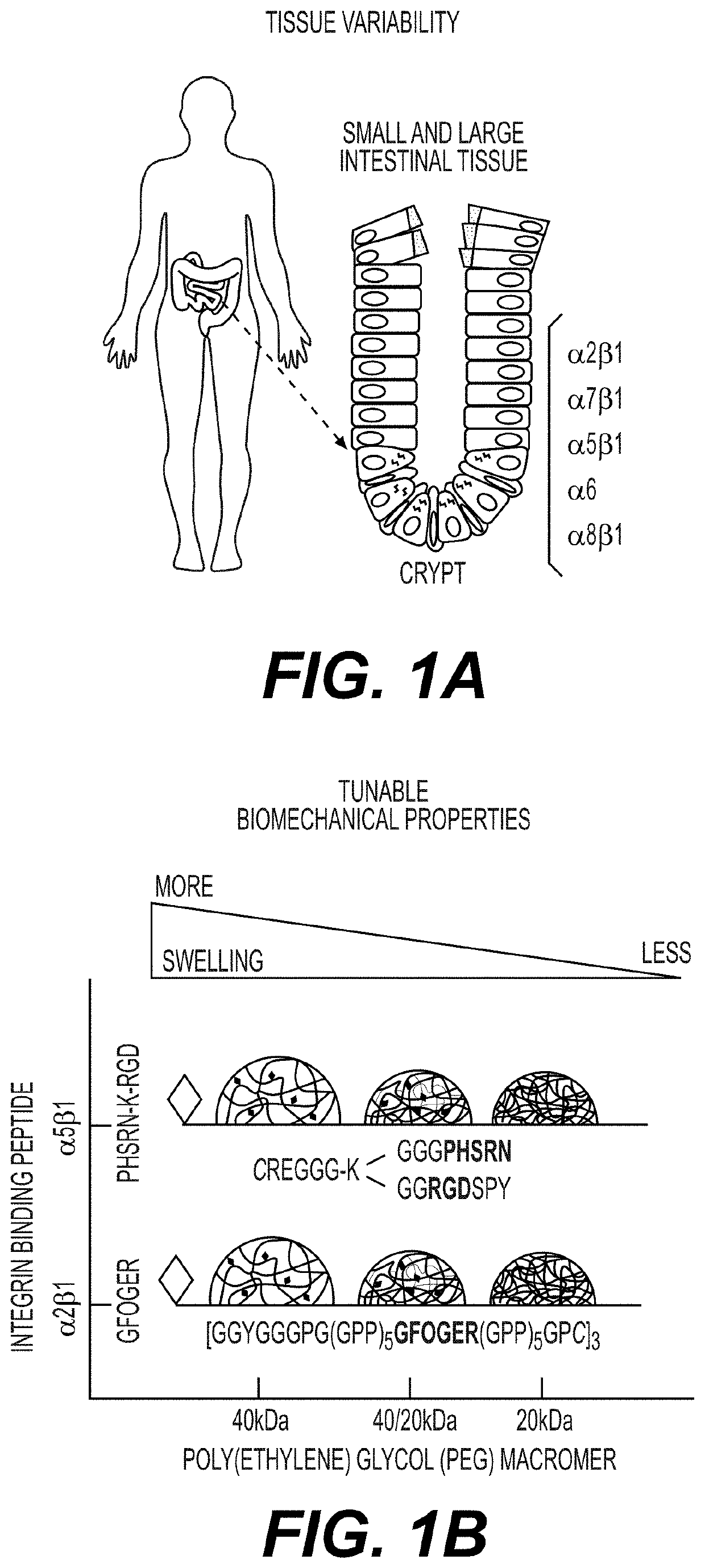
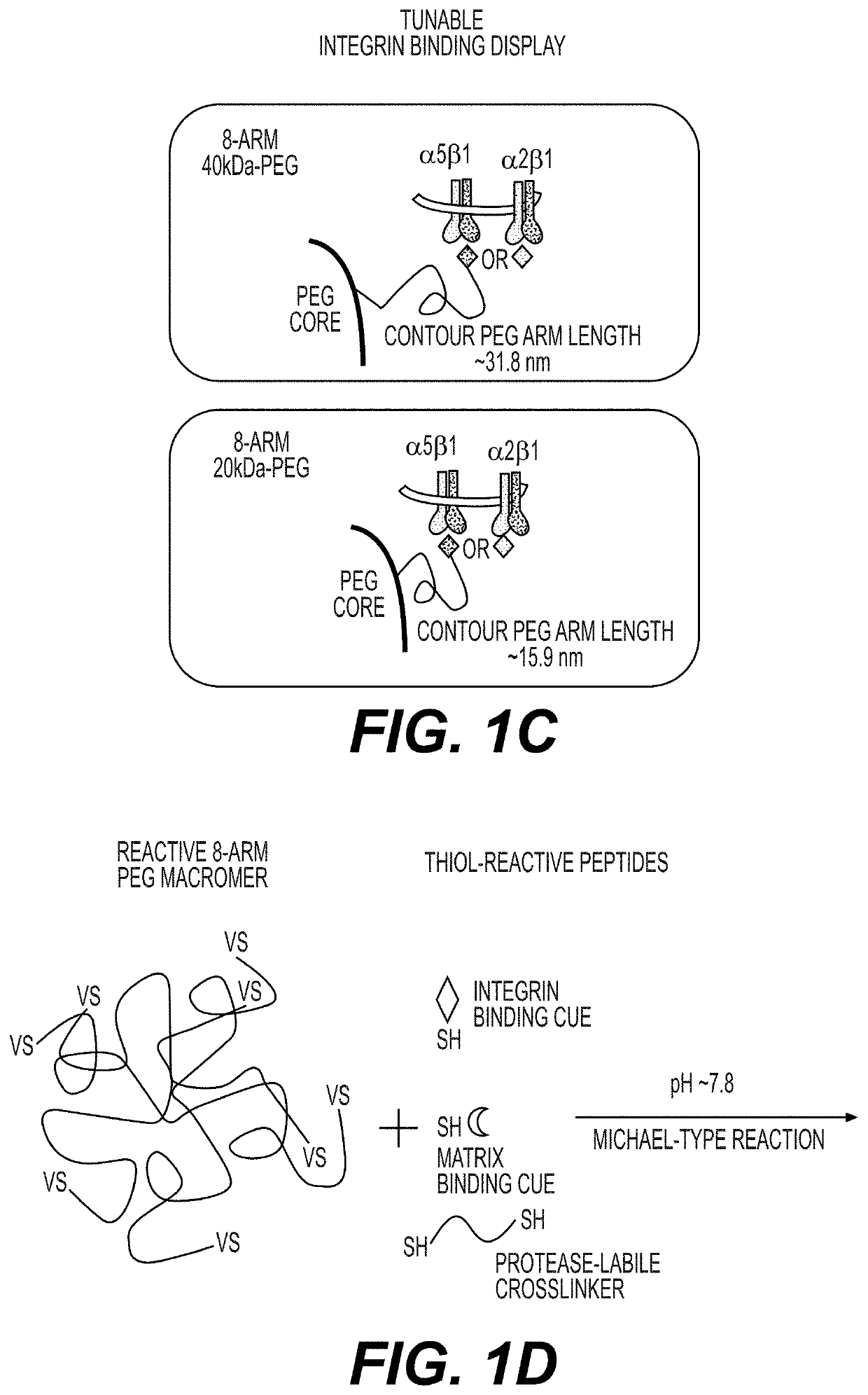
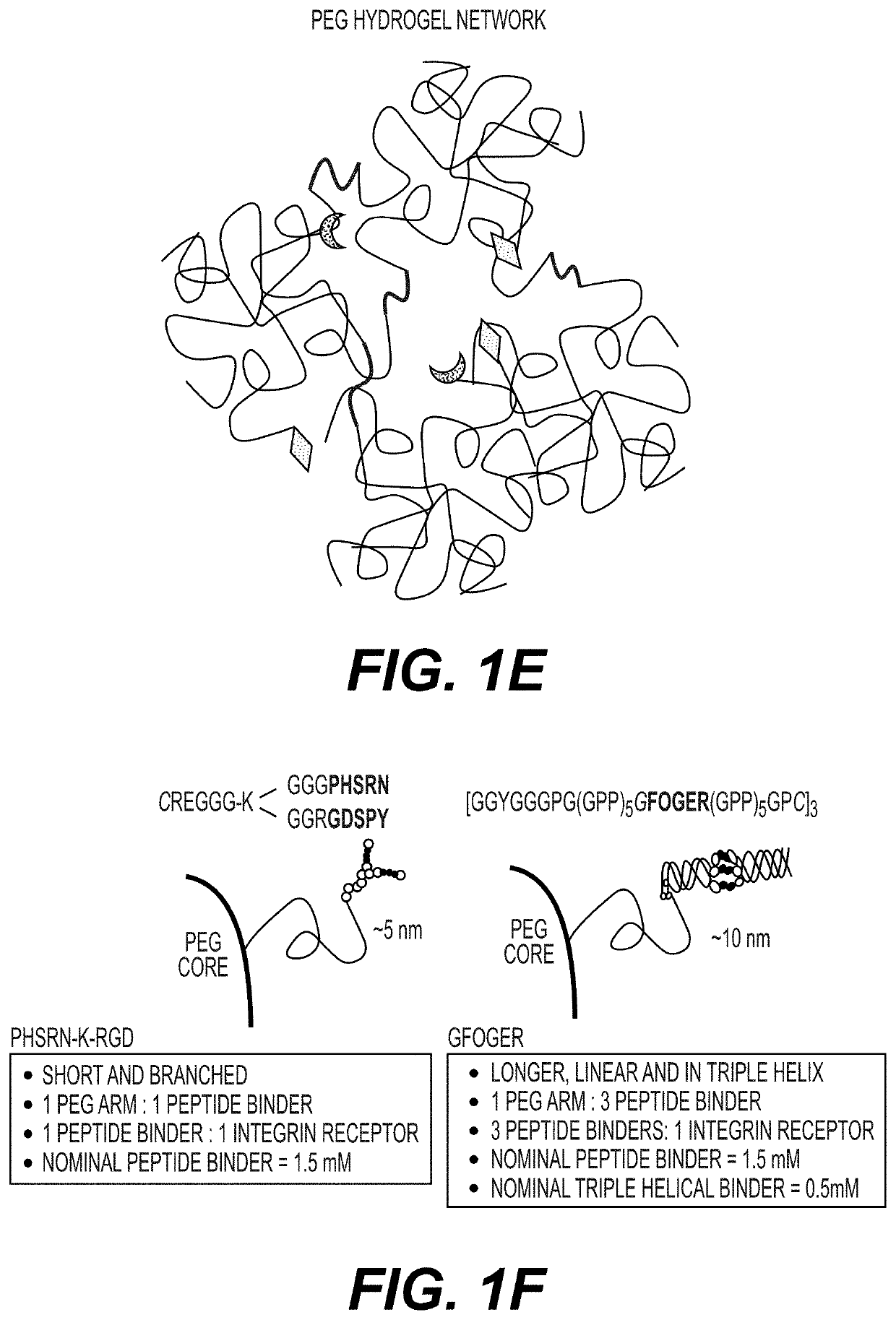
Synthetic hydrogels for organogenesis
PendingUS20210087534A1Gastrointestinal cellsArtificial cell constructsCell-Extracellular MatrixHuman cell
Synthetic hydrogels for organogenesis support organogenesis from mammalian cells, including human cells. The synthetic hydrogels typically include a network of crosslinked branched biodegradable polymers. A portion of the branches of the branched biodegradable polymers are linked to binders which are generally synthetic peptides for cell and extracellular matrix attachment. The hydrogels may include an inhibitor of apoptosis. The synthetic hydrogels with the synthetic binders typically do not interfere with cellular, proteomic, genetic, and / or transcriptome analyses of organoids formed in the hydrogel. The synthetic hydrogels may be subject to on-demand dissolution to provide intact organoids substantially free of hydrogel polymers. Also provided are methods of making the synthetic hydrogels and methods of using the synthetic hydrogels for organogenesis.
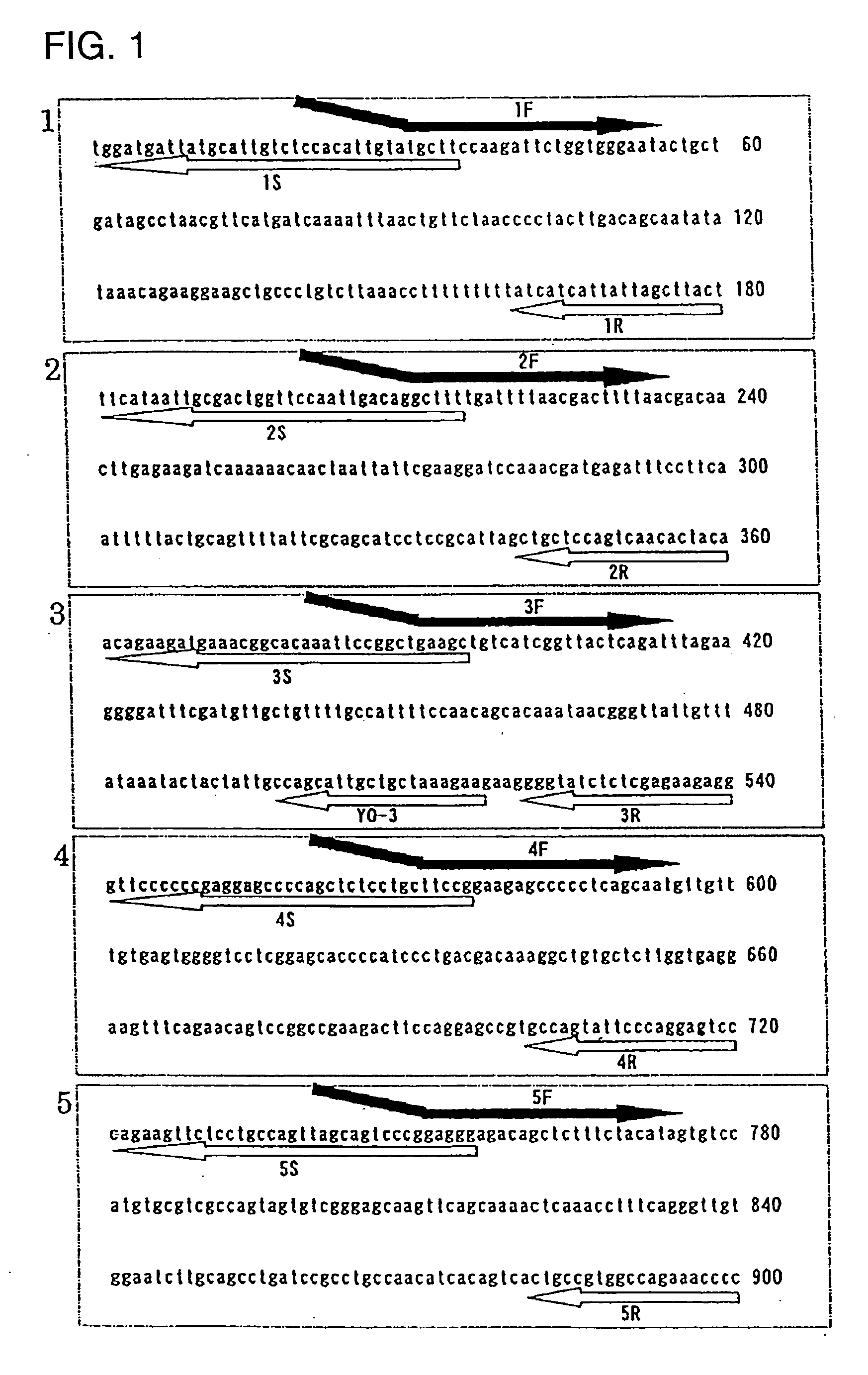
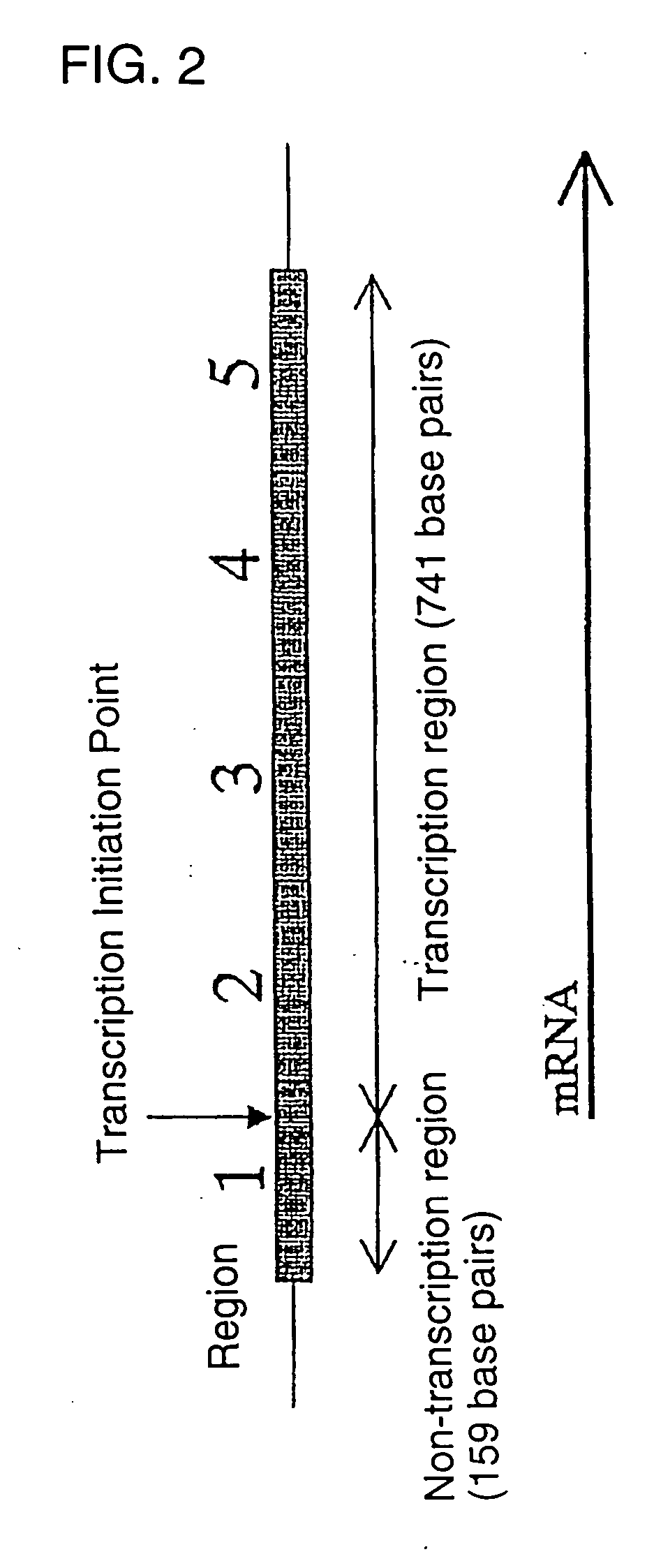
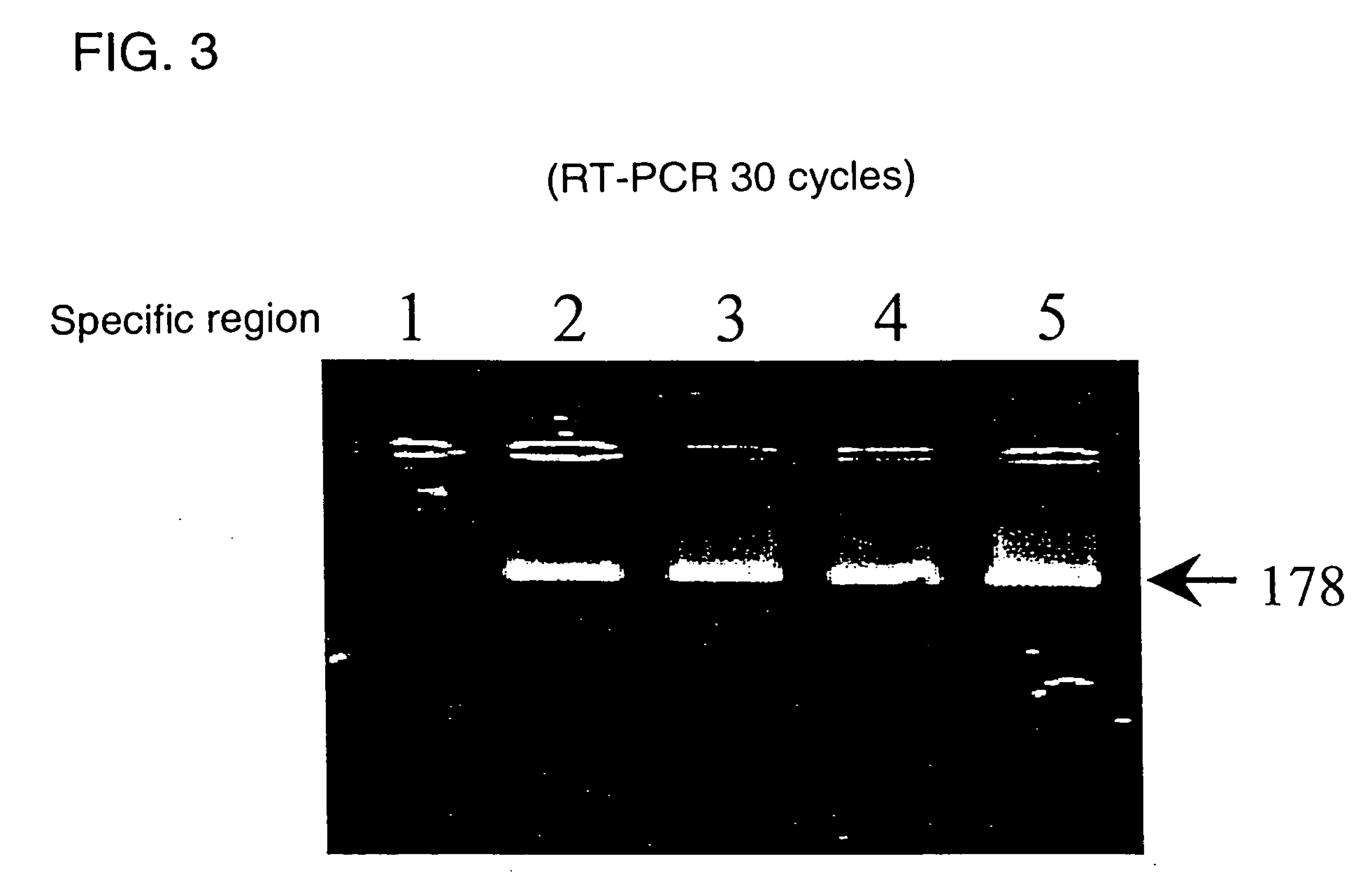
Novel genome analyzing method
InactiveUS20050064486A1Microbiological testing/measurementPeptide preparation methodsNucleotideNucleotide sequencing
A novel transcriptome analyzing method and to provide a gene found by this method and a protein encoded by the gene. A method for determining whether or not a continued arbitrary DNA sequence existing in the genome of an arbitrary biological species, in which the nucleotide sequence is already known but its possibility of being a gene expression region is unclear (specific region), is the specific region, which comprises detecting whether or not a nucleotide sequence that corresponds to the nucleotide sequence of the region is present in the RNA of the biological species, and a method for determining the gene expression region in an arbitrary region on a genome or the entire genome, which comprises repeatedly carrying out the above method.
Owner:TOSOH CORP
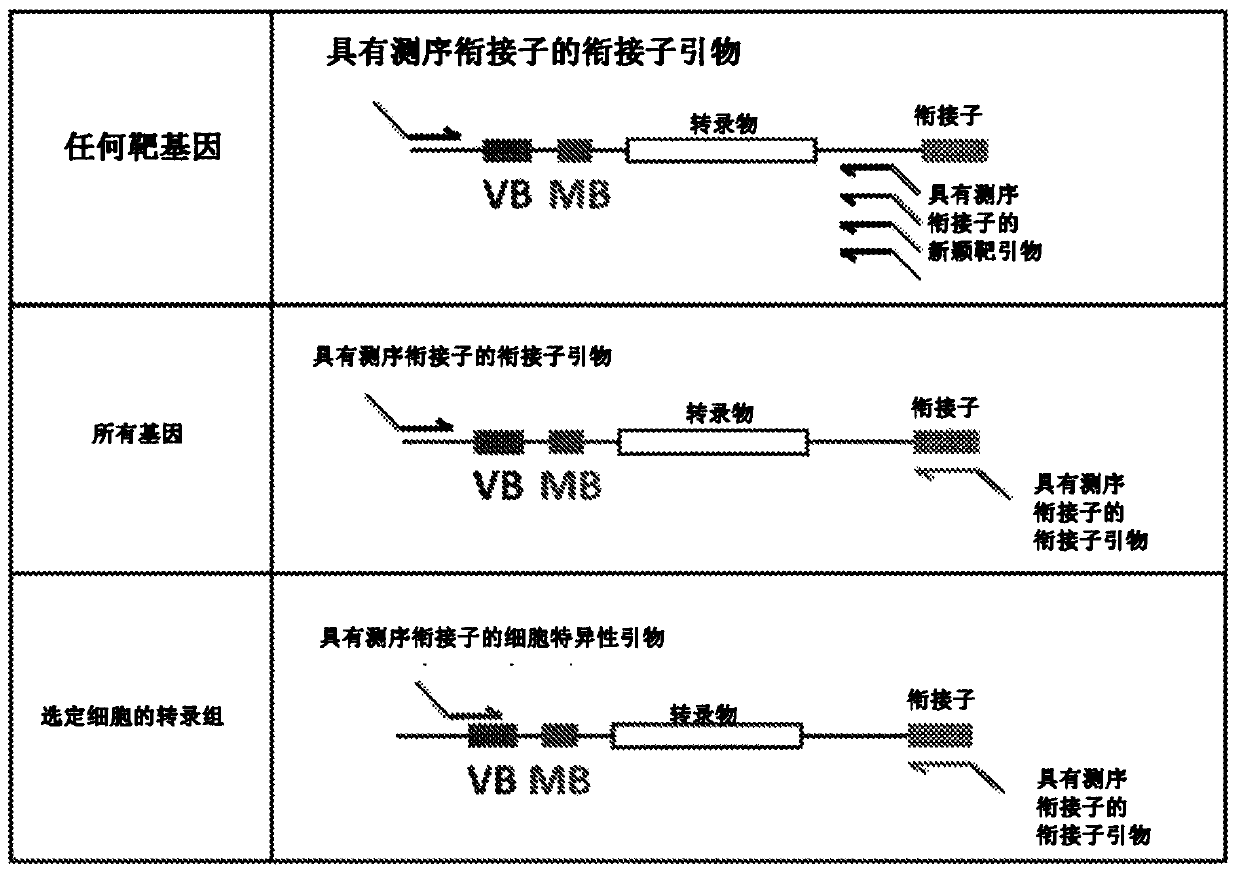
High-throughput polynucleotide library sequencing and transcriptome analysis
Provided herein are methods for target gene sequencing and single cell barcoding in conjunction with analysis of gene expression in single cells. In some embodiments, the target gene is an immune molecule, such as an antibody or TCR. In some embodiments, the methods can be used to carry out transcriptome sequencing, e.g., RNA sequencing, to capture transcriptome of single cells paired with full receptor immune receptor sequences such that information about the immune repertoire and transcriptome of a cell can be determined. Also provided are polynucleotide libraries for use in carrying out transcriptome analysis and immune molecule, e.g., antibody or TCR, sequencing.
Owner:ABVITRO LLC
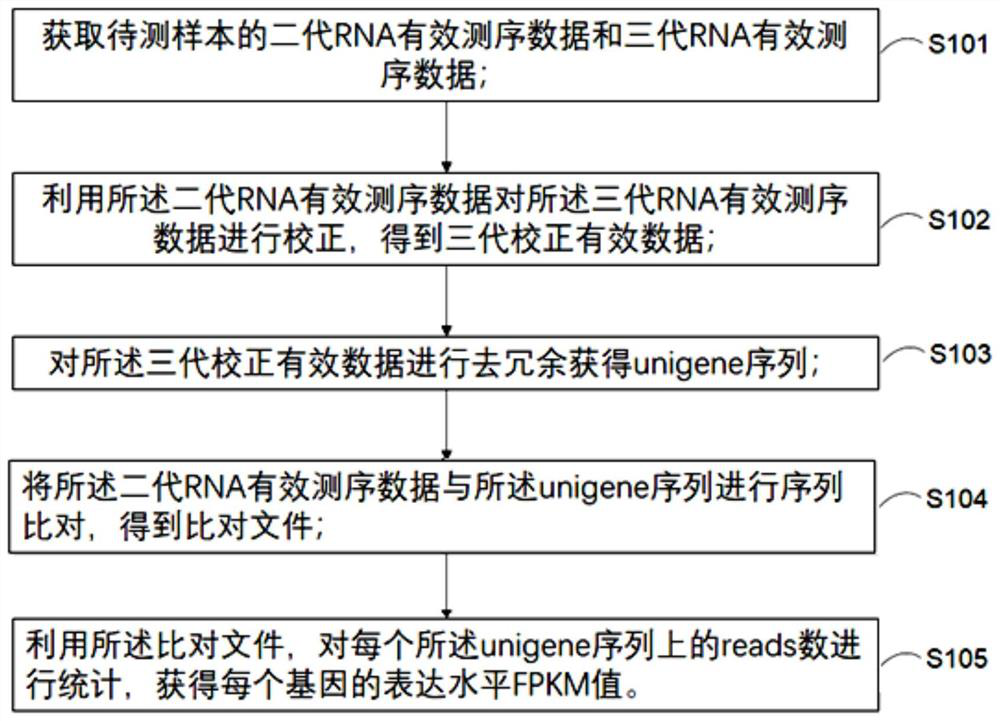
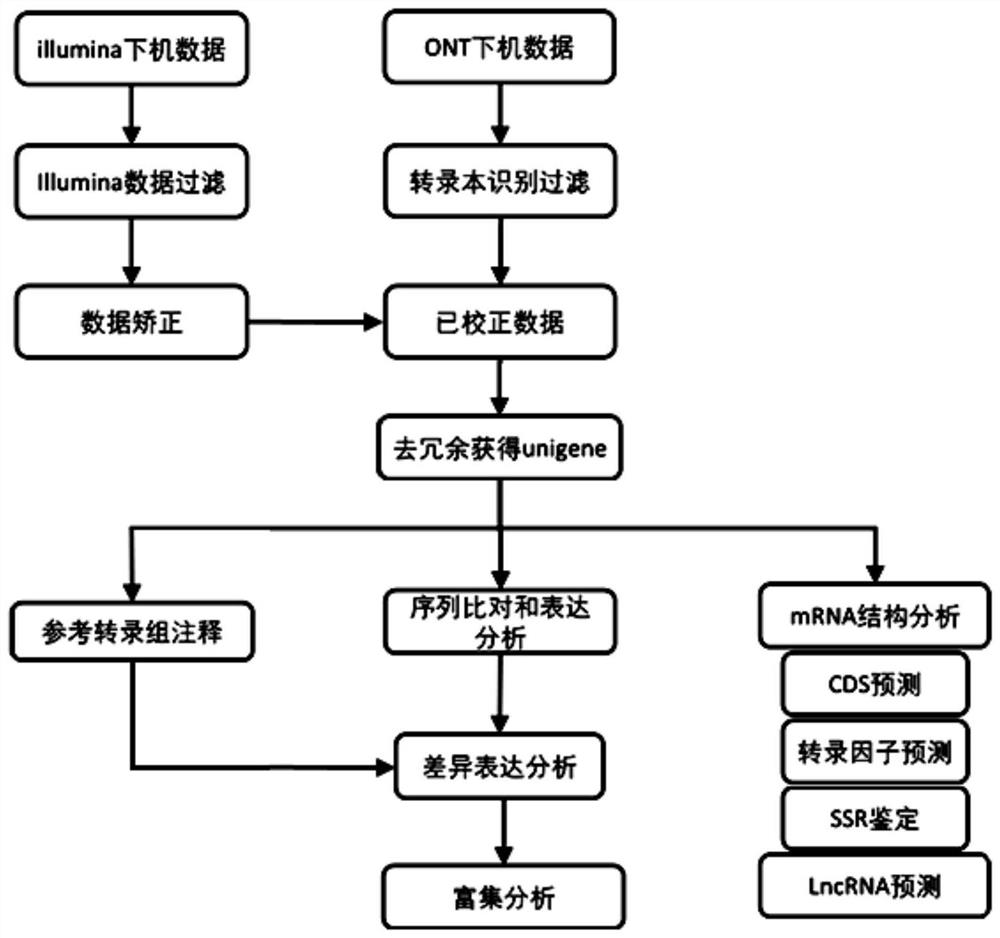
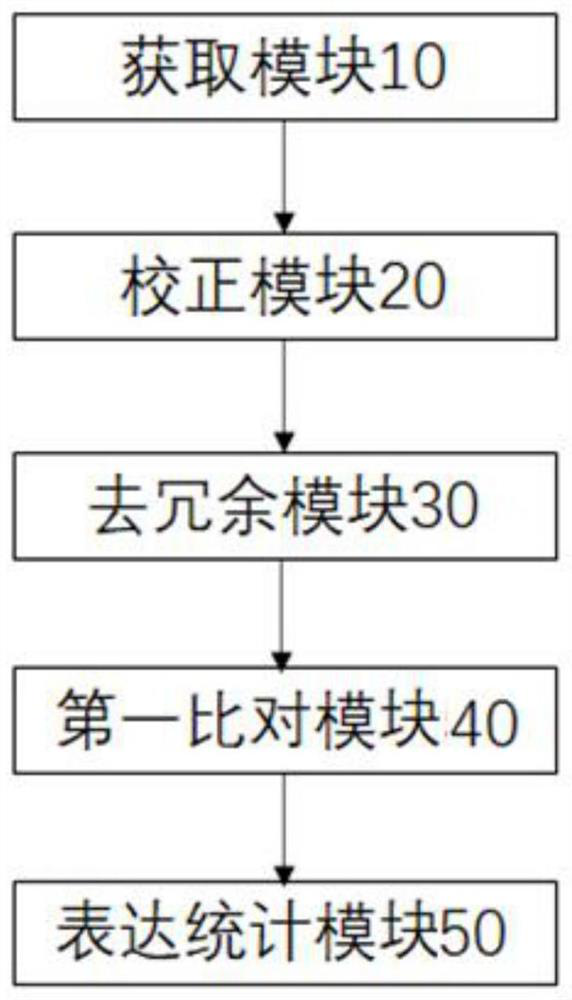
Transcriptome analysis method and system without reference genome sequence
The invention provides a transcriptome analysis method and system without a reference genome sequence. The analysis method comprises the following steps: acquiring second-generation RNA effective sequencing data and third-generation RNA effective sequencing data of a to-be-detected sample; correcting the third-generation RNA effective sequencing data by utilizing the second-generation RNA effective sequencing data; redundancy elimination is carried out on the valid data after the third-generation correction to obtain a unigene sequence; performing sequence comparison on the second-generation RNA effective sequencing data and the unigene sequences, and performing statistics on the reads number on each unigene sequence by utilizing a comparison file to obtain an expression level FPKM value of each gene. The analysis method integrates the advantages of a second-generation sequencing technology and a third-generation sequencing technology, and solves the problem that no transcriptome of areference-free genome sequence is analyzed by utilizing third-generation sequencing data at present.
Owner:TIANJIN MODERN INNOVATIVE TCM TECH CO LTD
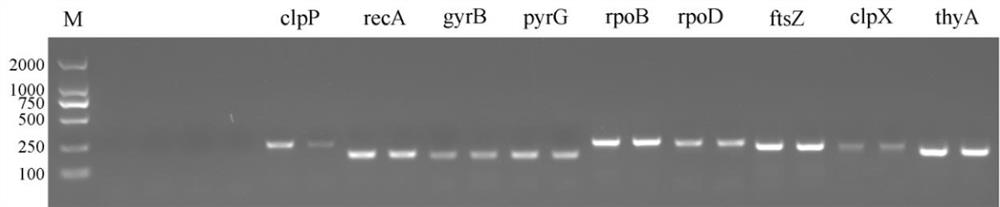
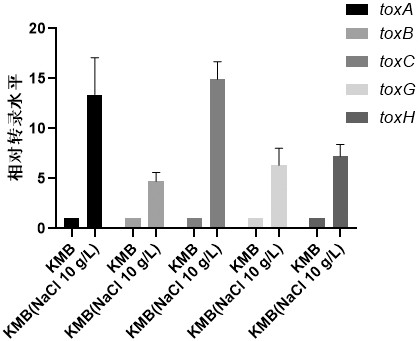
Screening and application of burkholderia gladioli real-time quantitative polymerase chain reaction (PCR) reference genes and primers thereof
ActiveCN111705149AImprove stabilityWide adaptabilityMicrobiological testing/measurementMicroorganism based processesReference genesCandidate Gene Association Study
The invention discloses screening and application of burkholderia gladioli real-time quantitative polymerase chain reaction (qRT-PCR) reference genes and primers thereof. According to obtained transcriptome analysis results and reported reference genes, 12 candidate reference genes with relatively stable expression are preliminarily screened, and gene names are shown as follows: atpD, clpP, clpX,ftsZ, gyrB, lpxC, pyrG, recA, rpoB , rpoD, thyA and 16S; for the candidate genes, amplification primers are designed; reference genes under different culture conditions (temperature, initial pH valueof medium, culture time, treatment with NaCl different in concentration) of burkholderia gladioli are screened; geNorm, NormFinder and Bestkeeper software is employed to analyze qRT-PCR results; and finally, the reference genes with most stable expression under different culture conditions and NaCl treatment condition are obtained by screening. The invention is beneficial to the stability and reliability of analysis and study of gene expression of the burkholderia gladioli under different conditions.
Owner:INST OF BOTANY JIANGSU PROVINCE & CHINESE ACADEMY OF SCI
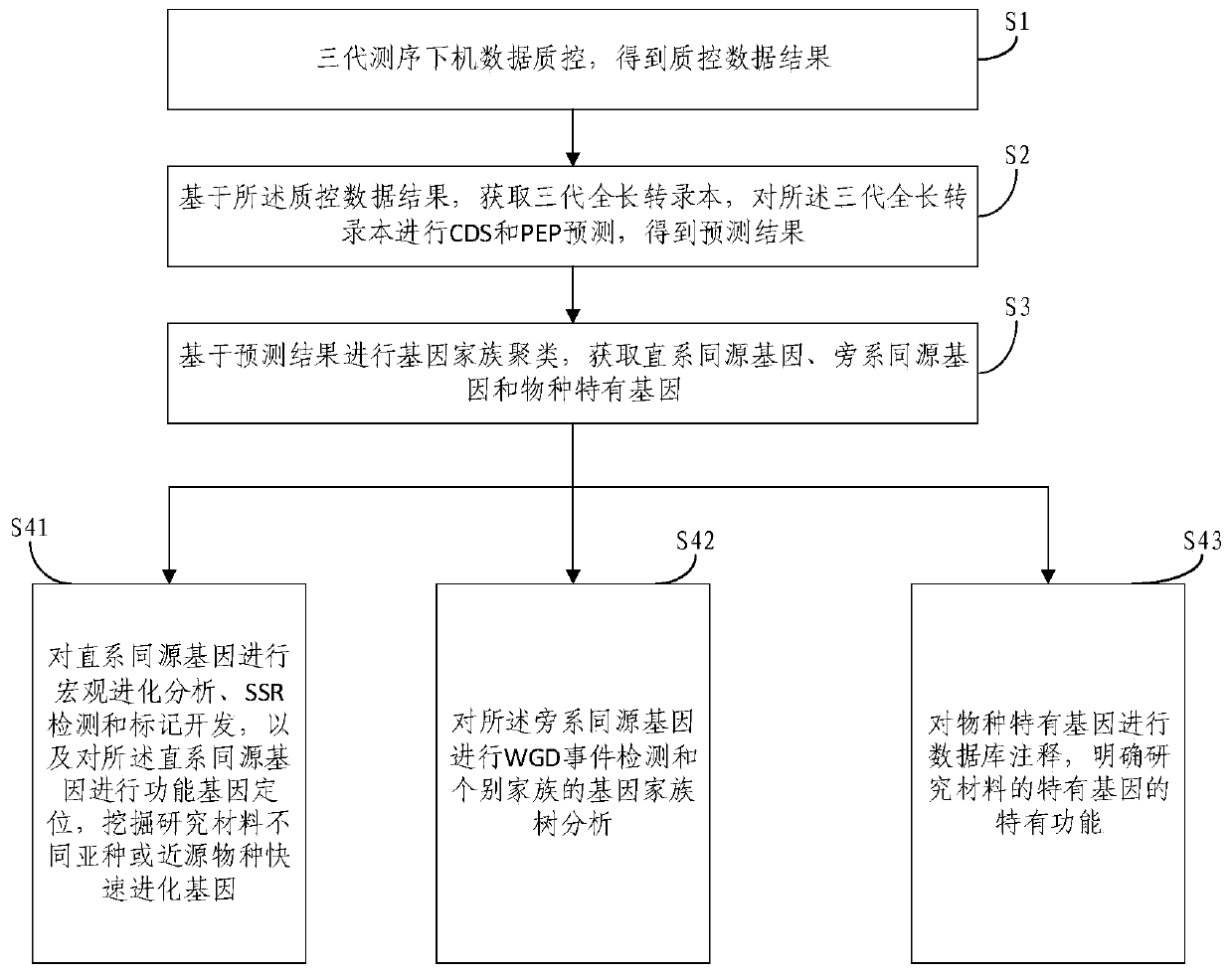
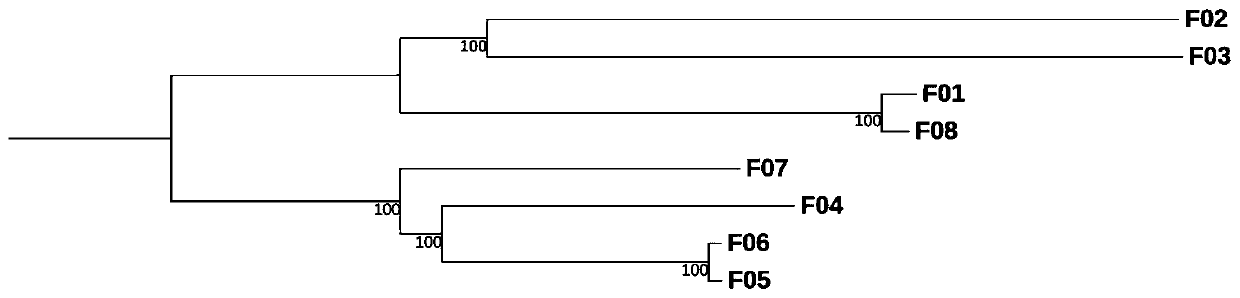
Analysis method for multi-sample-size comparative transcriptomes based on third-generation sequencing detection
An embodiment of the invention provides an analysis method for multi-sample-size comparative transcriptomes based on third-generation sequencing detection. The method comprises the following steps: carrying out quality control on data obtained after third-generation sequencing, acquiring a third-generation full-length transcript, carrying out CDS and PEP prediction, carrying out gene family clustering based on prediction results, and acquiring orthologous genes, paralogous genes and species-specific genes; carrying out macroscopic evolution analysis, SSR detection and marker development on theorthologous genes, carrying out functional gene localization, and excavating tachytelic evolution genes of different subspecies or near-source species of a researched material; carrying out WGD eventdetection and gene family tree analysis of individual families on the paralogous genes; and subjecting the species-specific genes to database annotation to clearly learn about the specific functionsof the specific genes of the researched material. The embodiment of the invention provides a set of perfect scientific research method for bioinformatics research on the whole so as to realize processing of multi-sample-size comparative transcriptome data.
Owner:BIOMARKER TECH
Characteristic analysis method and classification of pharmaceutical components by using transcriptomes
InactiveUS20190325991A1Accurate predictionCompounds screening/testingHeating or cooling apparatusAdjuvantOrganism
The present invention provides a novel method for the classification of adjuvants. In one embodiment, the present invention provides a method for generating organ transcriptome profiles for adjuvants, said method comprising: (A) a step for obtaining expression data by performing transcriptome analysis for at least one organ of a target organism by using at least two adjuvants; (B) a step for clustering the adjuvants with respect to the expression data; and (C) a step for generating the organ transcriptome profile for the adjuvants on the basis of the clustering.
Owner:NAT INST OF BIOMEDICAL INNOVATION HEALTH & NUTRITION
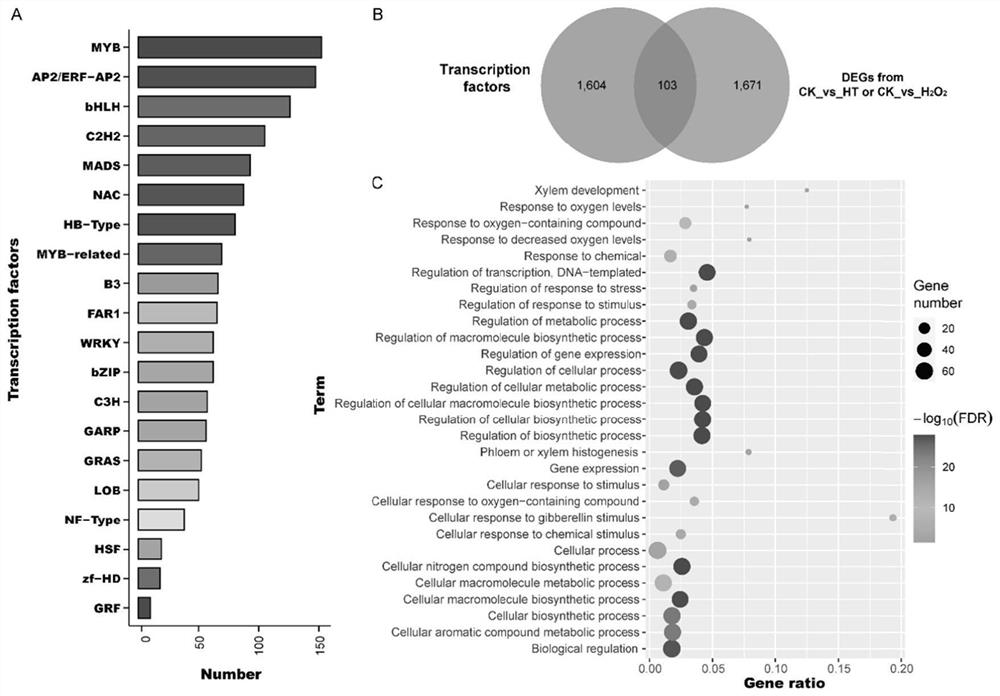
Method for screening transcription factors related to postharvest softening of grape fruits
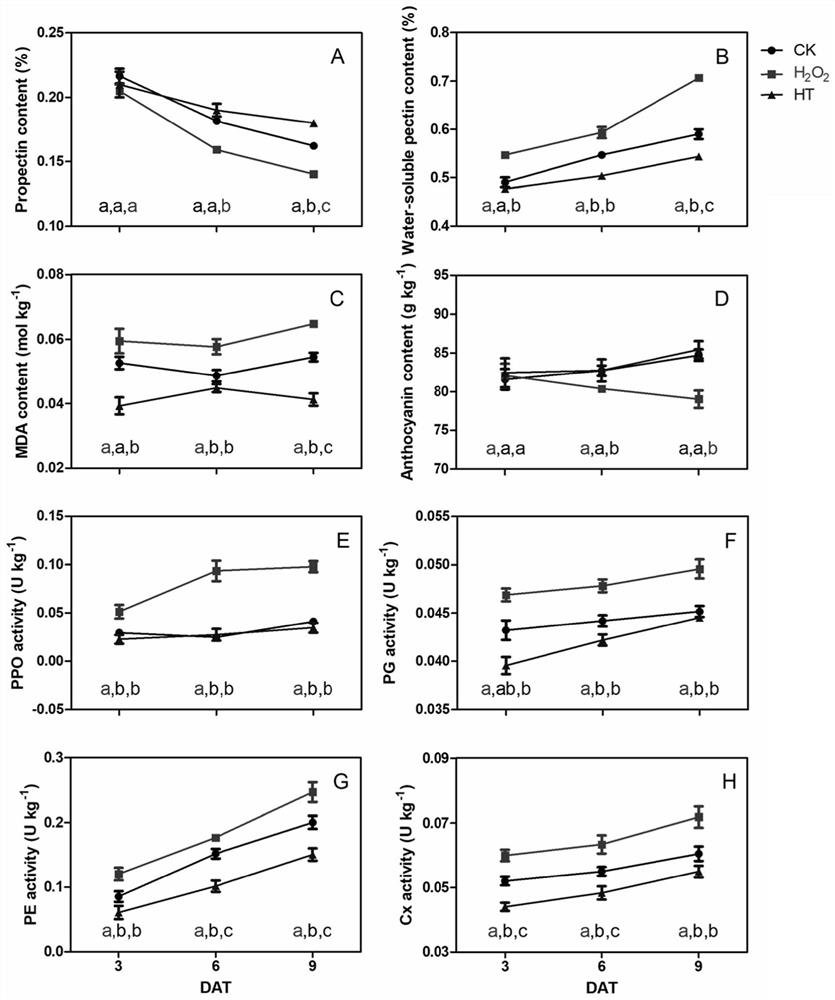
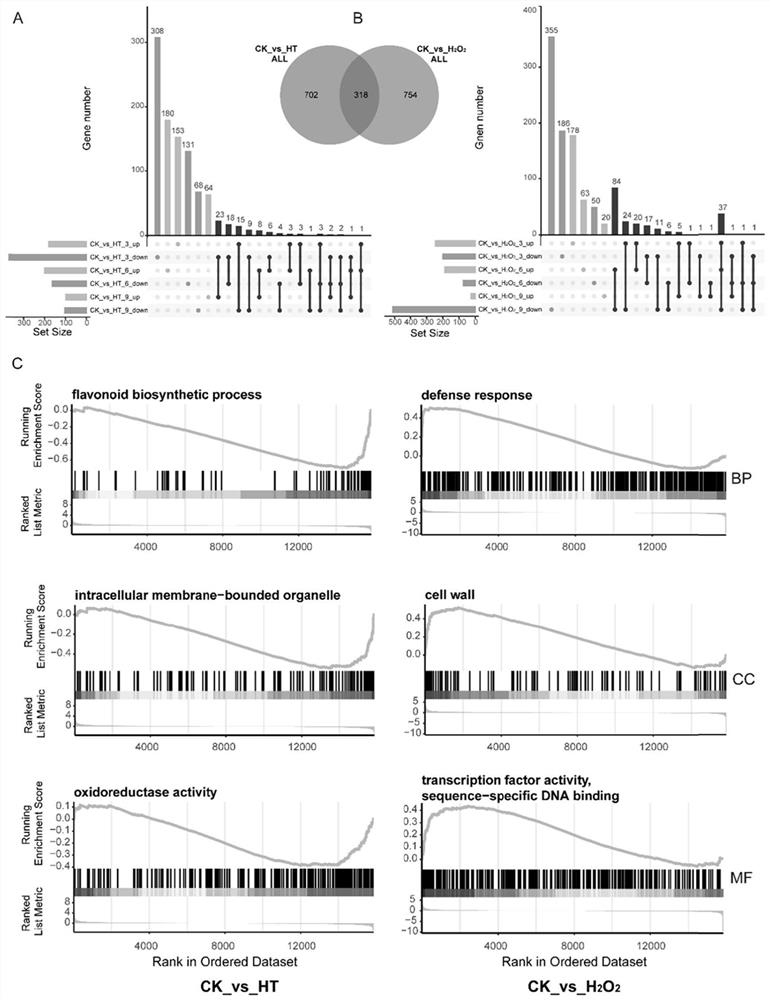
InactiveCN114107456AMicrobiological testing/measurementProteomicsBiotechnologyH2O2 - Hydrogen peroxide
The invention belongs to the technical field of high-throughput sequencing data analysis, and particularly relates to a method for screening transcription factors related to postharvest softening of grape fruits. According to the invention, hypotaurine (HT) and hydrogen peroxide (H2O2) treatment is carried out on harvested 'Kyoho' grapes, through transcriptome sequencing and weighted gene co-expression network analysis (WGCNA), correlation analysis is carried out on differential expression genes obtained through transcriptome analysis and pectin content change, a differential expression gene module closely related to the pectin content change is screened out, and the specific value of the differential expression genes and the specific value of the pectin content change are obtained. And excavating differential expression transcription factors in the softening process of the picked grape fruits. According to the method, the softening character related transcription factors can be effectively screened, and a theoretical reference is provided for researching the softening occurrence mechanism of the picked grape fruits.
Owner:HENAN UNIV OF SCI & TECH
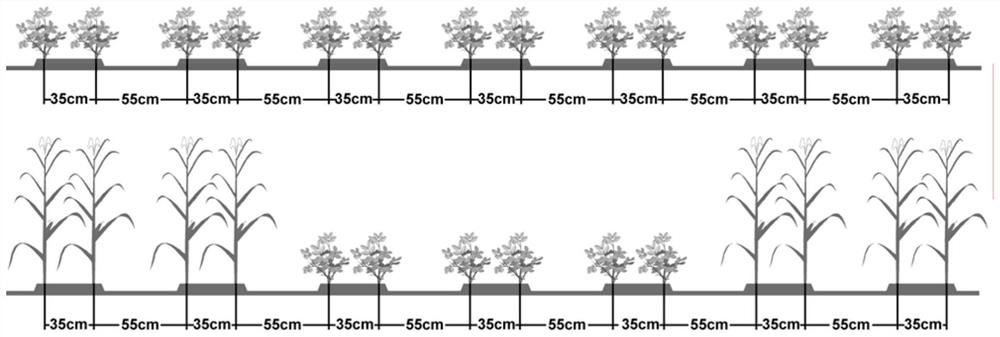
Comparative transcriptome analysis method for peanut leaf gene differential expression under intercropping corn
ActiveCN113337633AReduce adverse effectsConsistent fold changeMicrobiological testing/measurementSequence analysisBiotechnologyMonocropping
The invention relates to the technical field of biology, and particularly relates to a comparative transcriptome analysis method for peanut leaf gene differential expression under intercropping corn. The method comprises the steps that an RNA-seq technology is utilized for comparing and analyzing differential expression genes of peanut functional leaves in the pod expansion period under peanut single cropping and intercropping corn stress; differential genes related to metabolic pathways are taken as main genes, important genes and key metabolic pathways formed by peanut pod yield under intercropping corn stress are excavated, and a theoretical basis is provided for reducing adverse effects of intercropping corn stress on peanut yield by means of a directional chemical regulation and control technology; and the relative expression level of candidate differential expression genes is verified through qRT-PCR detection, and finally the qRT-PCR detection result is roughly consistent with multiple changes in transcriptome sequencing analysis. The transcriptome sequencing analysis method is accurate, feasible, effective and simple.
Owner:GUANGXI ZHUANG AUTONOMOUS REGION ACAD OF AGRI SCI
Primer pair for identifying carrot petal cytoplasmic male sterility and application thereof
ActiveCN114150084ATroubleshoot identification issuesSolve the problem of early removal of impuritiesMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyTotal rna
The invention discloses a primer pair for identifying carrot petal cytoplasmic male sterility and application thereof, the primer pair is designed according to an Indel molecular marker screened by transcriptome analysis, and the application is that the primer pair is adopted to identify carrot petal cytoplasmic male sterility. According to the application, transcriptome sequencing analysis is carried out on total RNA (Ribonucleic Acid) of a group of stable carrot petal cytoplasmic male sterile lines and maintainer lines thereof, comparison with a published carrot reference mitochondrial genome is carried out, Indel loci with differences between the sterile lines and the maintainer lines are screened out, and primers are designed. Specific amplification verification is carried out between fertile plants and sterile plants of different varieties of carrots, the result shows that the primer pair can completely distinguish the fertility of the variety to be detected, the carrot variety with cytoplasmic male sterility can be rapidly screened out in the seedling stage, the accuracy rate is high, and the method can be used for removing abnormal plants in the breeding process. The method has important practical application value and market prospect.
Owner:天津市农业科学院
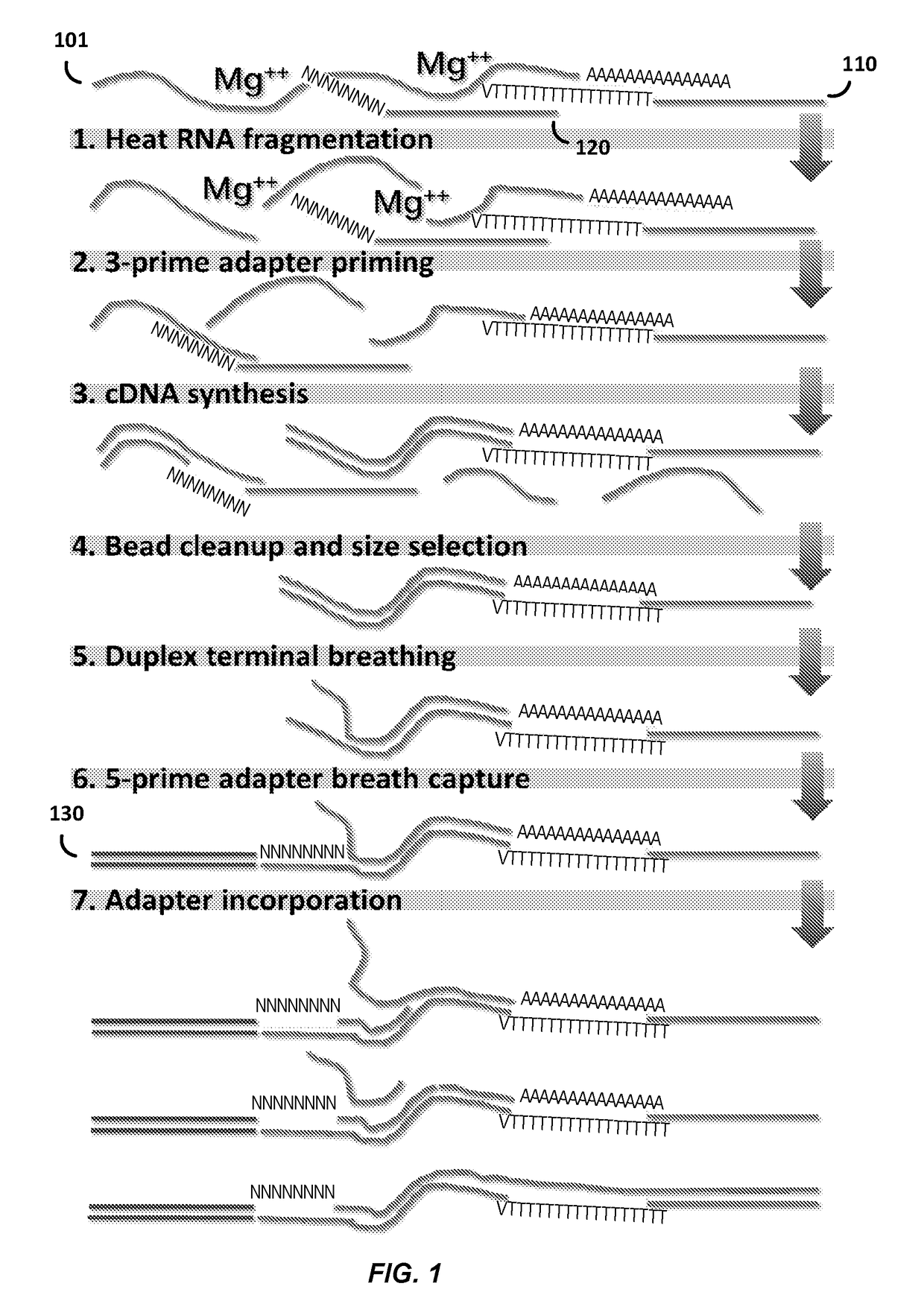
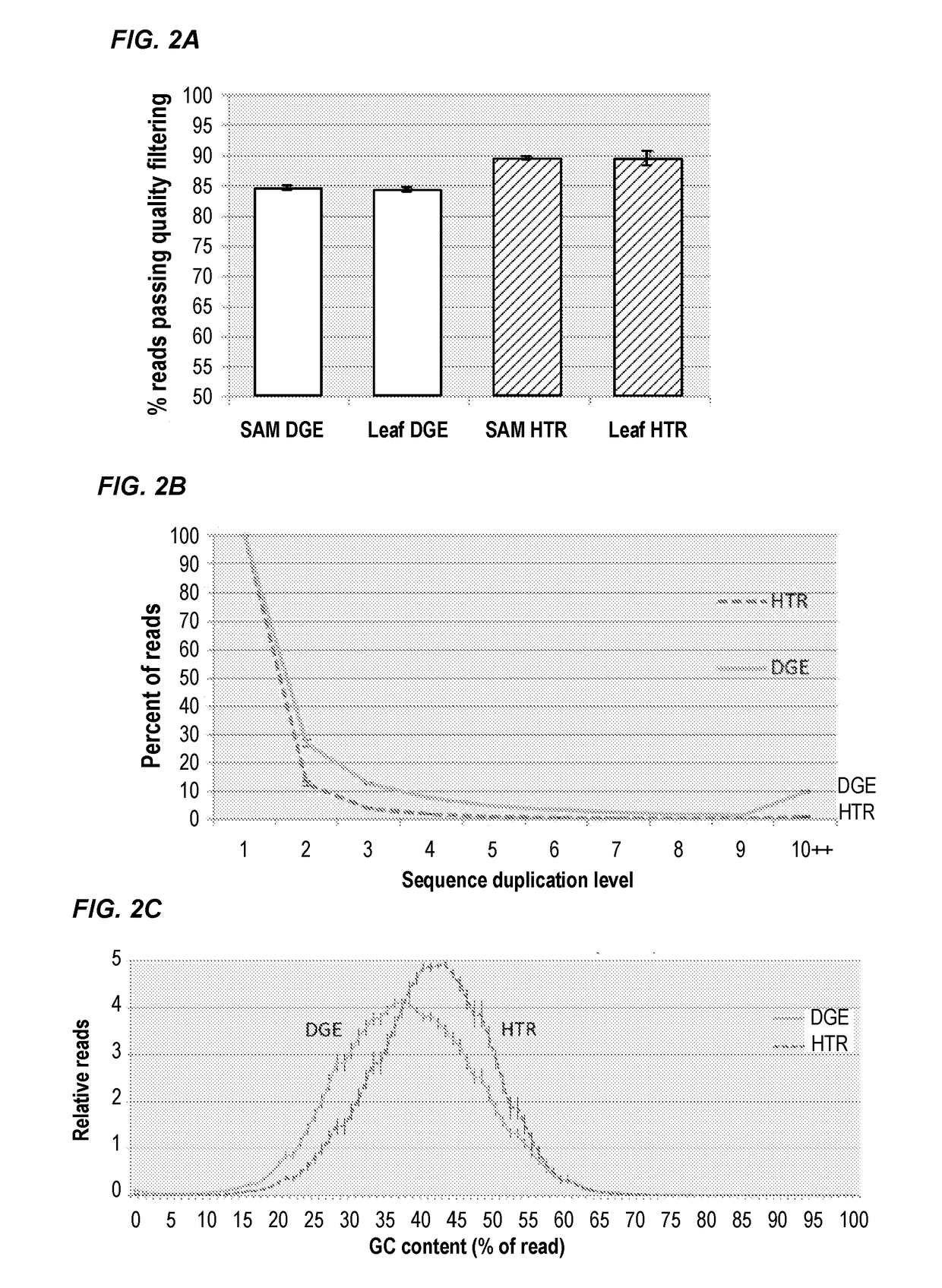
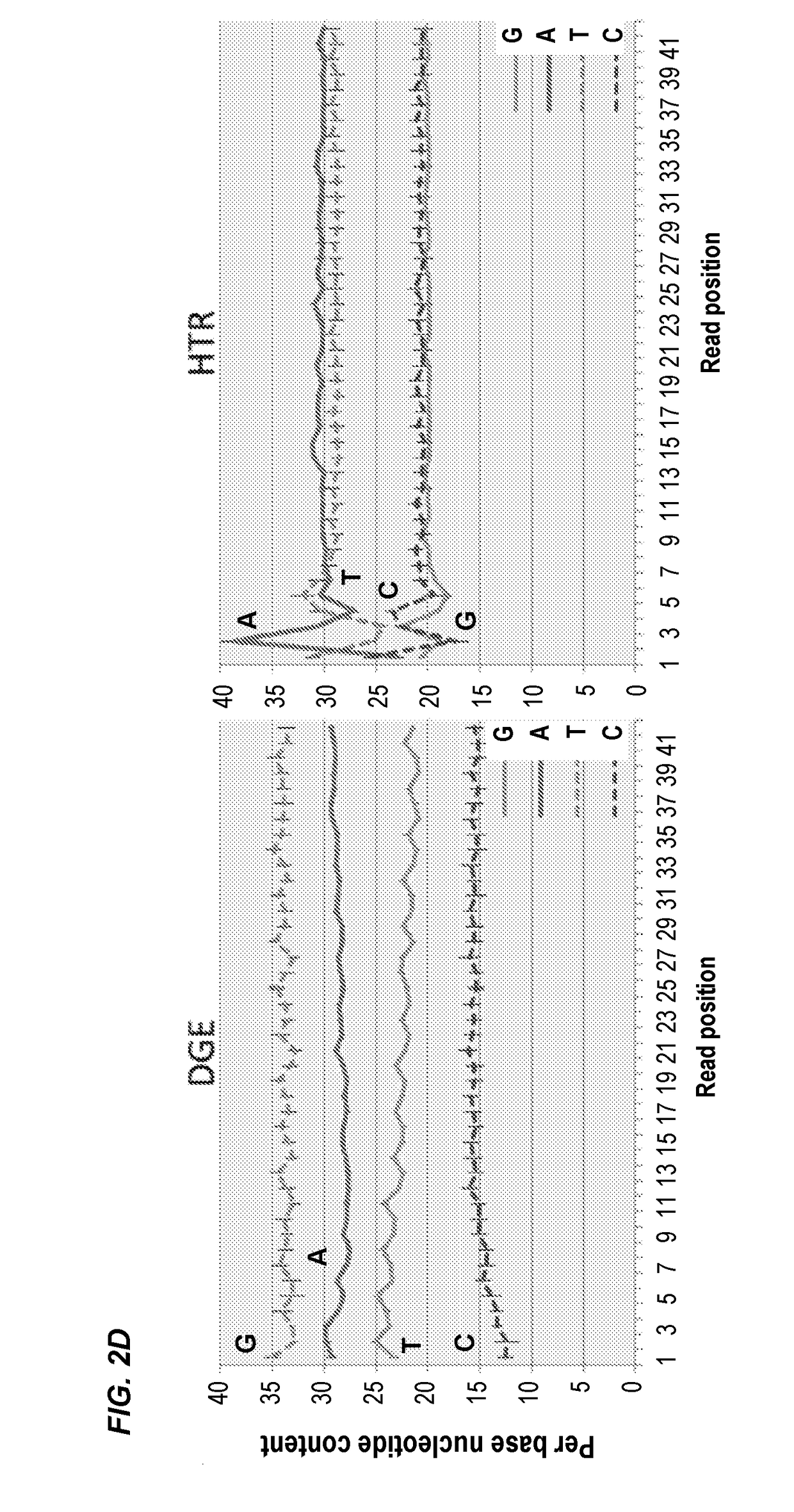
Compositions and methods for constructing strand specific cdna libraries
Provided herein are compositions, kits and methods for the production of strand-specific cDNA libraries. The compositions, kits and methods utilize properties of double stranded polynucleotides, such as RNA-cDNA duplexes to capture and incorporate a novel sequencing adapter. The methods are useful transcriptome profiling by massive parallel sequence, such as full-length RNA sequencing (RNA-Seq) and 3′ tag digital gene expression (DGE).
Owner:RGT UNIV OF CALIFORNIA
Selective extension in single cell whole transcriptome analysis
ActiveUS20200190564A1Microbiological testing/measurementDNA preparationCell biologyNUCLEIC ACID SUBSTANCE
Disclosed herein include methods and compositions for selectively amplifying and / or extending nucleic acid target molecules in a sample. The methods and compositions can, for example, reduce the amplification and / or extension of undesirable nucleic acid species in the sample, and / or allow selective removal of undesirable nucleic acid species in the sample.
Owner:BECTON DICKINSON & CO
A method for single-cell RNA reverse transcription and library construction
ActiveCN108103055BAvoid pollutionReduce inputMicrobiological testing/measurementLibrary creationCDNA libraryTotal rna
Owner:XUKANG MEDICAL SCI & TECH (SUZHOU) CO LTD
Method for predicting the response of antipsychotic drugs
PendingUS20220290237A1Promote resultsIncrease powerOrganic active ingredientsMicrobiological testing/measurementVitamin B6 synthesisAmisulpride
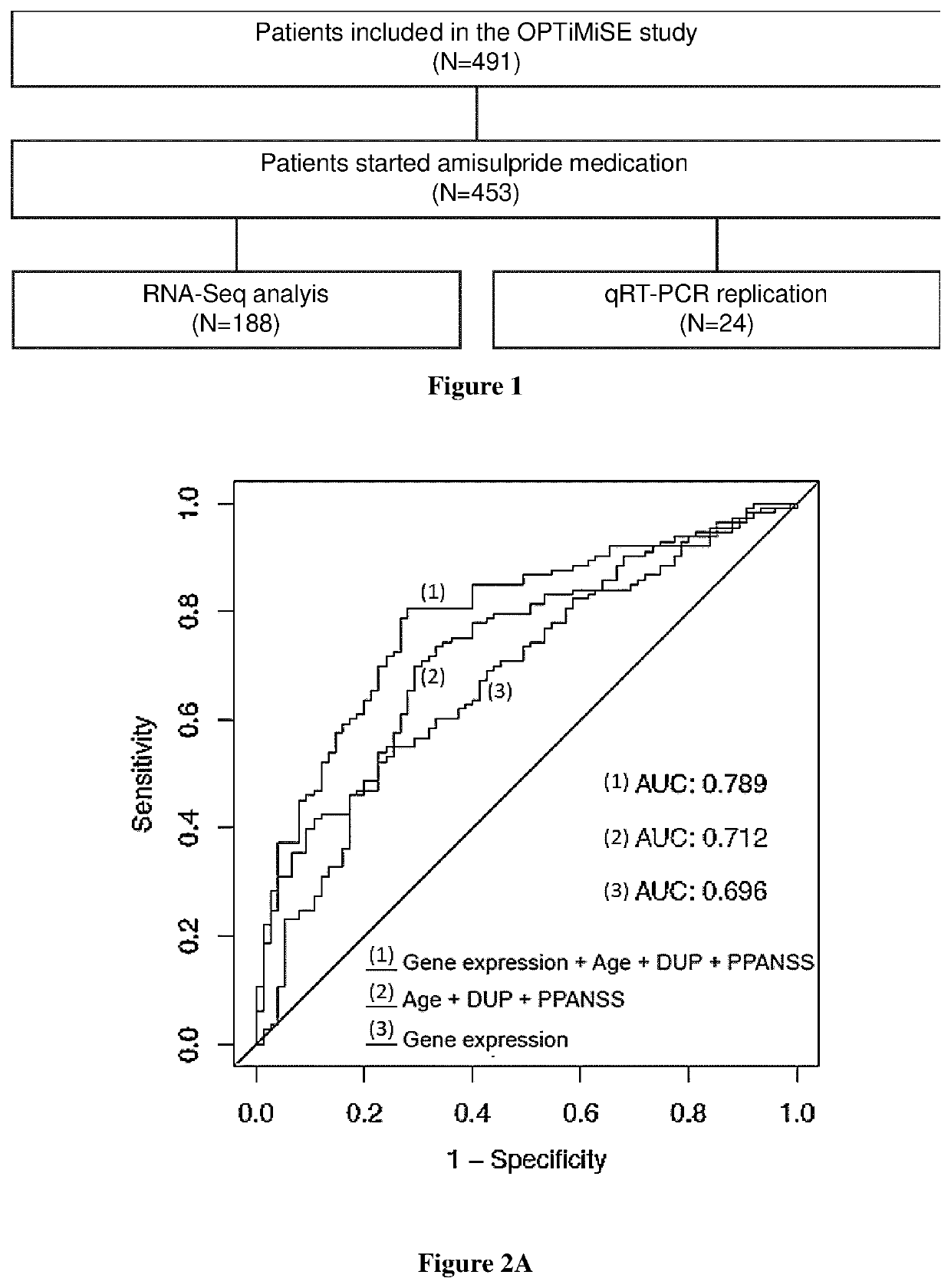
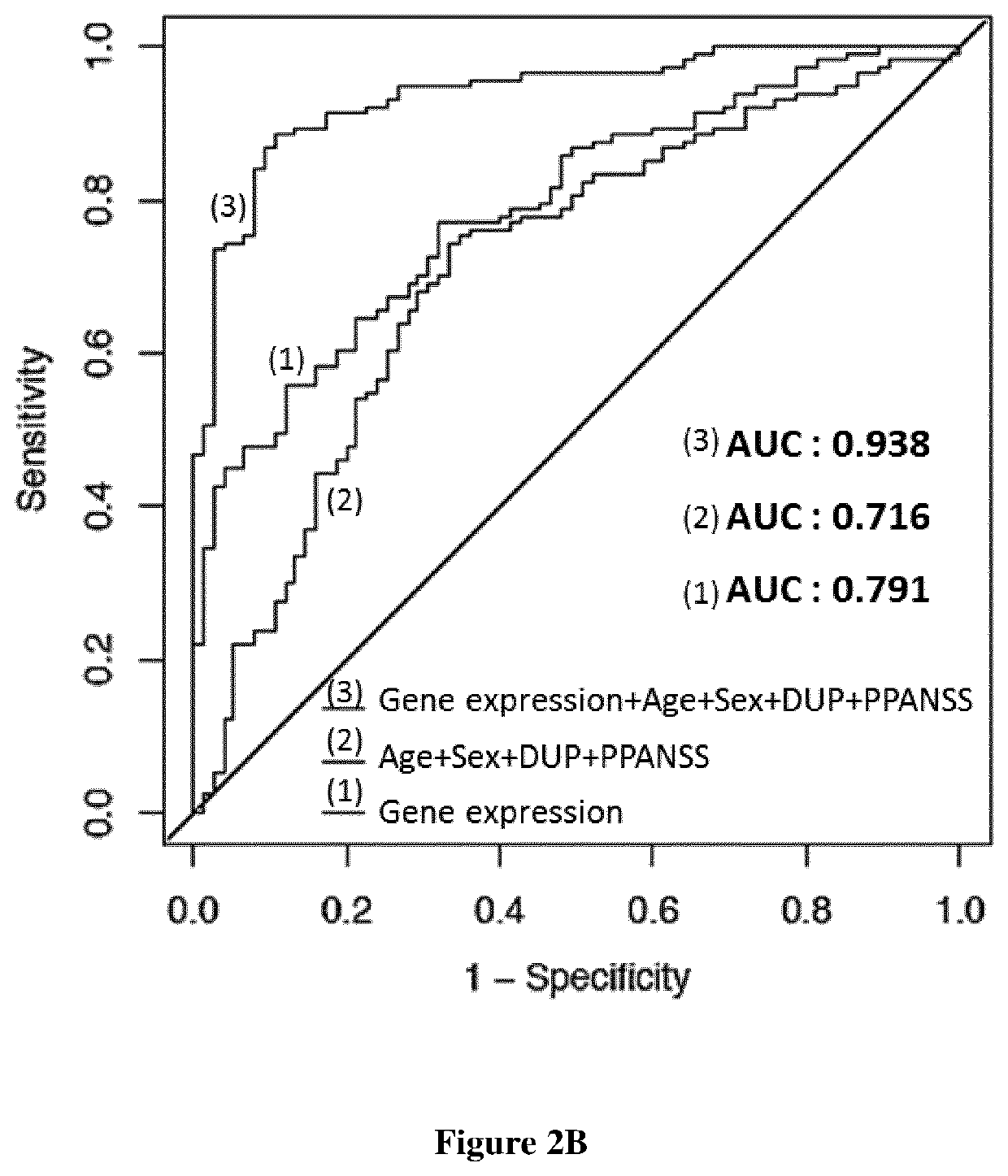
A fundamental shortcoming in the current treatment of schizophrenia is the lack of valid criteria to predict who will respond to antipsychotic treatment. The identification of blood-based biological markers of the therapeutic response would enable clinicians to identify the subgroup of patients in whom conventional antipsychotic treatment is ineffective and offer alternative treatments. As part of the Optimization of Treatment and Management of Schizophrenia in Europe (OPTiMiSE) programme, the inventors conducted a transcriptome analysis on 188 subjects with first episode psychosis, all of whom were subsequently treated with amisulpride for 4 weeks. They identify 32 genes for which the expression changed after treatment in good responders only. Among these genes, the expression of ALPL, a gene involved in vitamin B6 metabolism, as well as CA4, DGTA2, DHRS13, HOMER3 and WLS showed a significant difference in expression level between good and poor responders before starting treatment, allowing to predict treatment outcome with a predictive value of 93.8% when combined with clinical features Collectively, these findings identified new mechanisms to explain symptom improvement after amisulpride medication and highlight the potential of combining gene-expression profiling with clinical data to predict treatment response in first episode psychoses.
Owner:INST NAT DE LA SANTE & DE LA RECHERCHE MEDICALE (INSERM) +1
Novel single-celled spatial transcriptome technology for tissue
ActiveCN113322314AMicrobiological testing/measurementPreparing sample for investigationHigh fluxTissue sample
The invention provides a single-celled spatial transcriptome analysis method. Specifically, the invention provides a single-celled spatial transcriptome capable of detecting an undissociated state. According to the method provided by the invention, single cells can be collected in a high flux manner on the basis of reserving spatial position information of cells in tissue, quantified cellular transcriptome data with high sensitivity and high repeatability are obtained through secondary high-flux sequencing, single-celled spatial transcriptome sequencing data of each target cell are mapped to corresponding spaces of a tissue sample based on three-dimensional coordinate values of each target cell, and thus, a single-celled spatial transcriptome sequencing data set of the tissue sample is obtained. The method provided by the invention can be extensively applied to quantitative research on single-celled gene expression of normal and diseased tissue and provides a more efficient and accurate tool for researching states and functions of cells of the tissue.
Owner:SHANGHAI JIAO TONG UNIV
Whole transcriptome analysis in single cells
PendingUS20220177950A1Reduced stabilityMicrobiological testing/measurementDNA preparationSingle cell transcriptomeReverse transcriptase
The invention is a method of single cell transcriptome analysis. The method comprises detecting multiple transcripts in each individual cell of the plurality of cells by barcoding the transcripts with a cell-specific compound barcode formed using a DNA polymerase and a terminal transferase, optionally in a single enzyme such as a reverse transcriptase.
Owner:ROCHE SEQUENCING SOLUTIONS INC
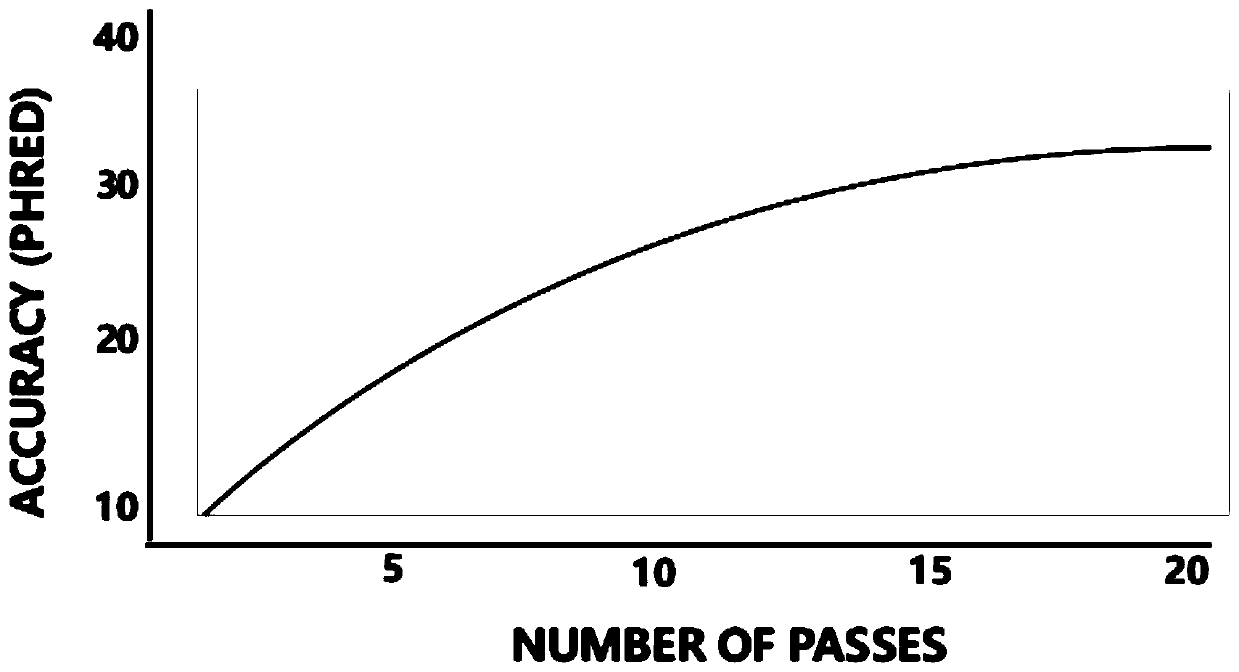
Third-generation full-length transcriptome sequencing result analysis method suitable for Sequel sequencing
The invention discloses a third-generation full-length transcriptome analysis method suitable for a Sequel sequencing platform, which is characterized by comprising the following steps: 1, a sequencing data filtering step; 2, a sequencing data comparison step; 3, a transcript annotation step; 4, an ORF prediction step; 5, a transcript function annotation step; 6, a fusion gene analysis step; 7, aLncRNA prediction step; 8, variable shear analysis step; and 9, a variable polyadenylation analysis step. According to the method, the running speed is higher, annotation on the transcript is finer compared with common matchannot software, and the type of the transcript is more convenient to analyze.
Owner:南京派森诺基因科技有限公司
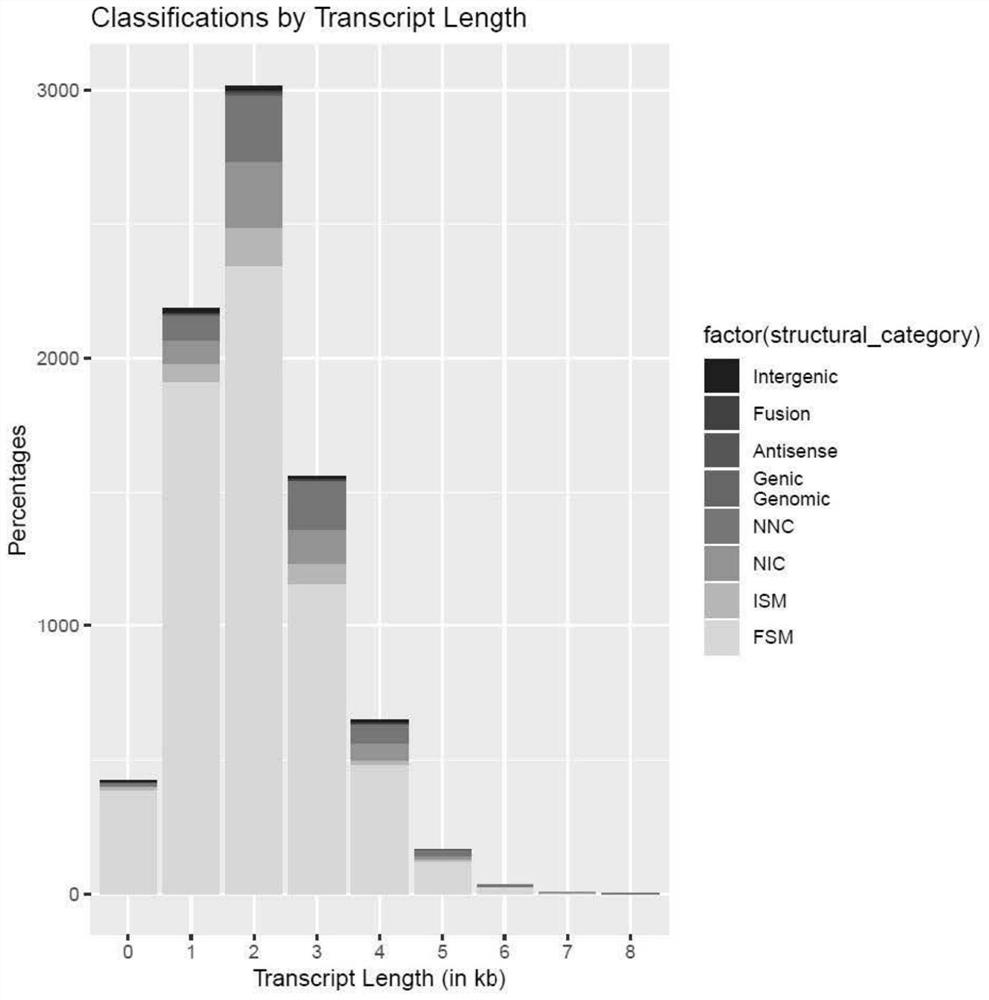
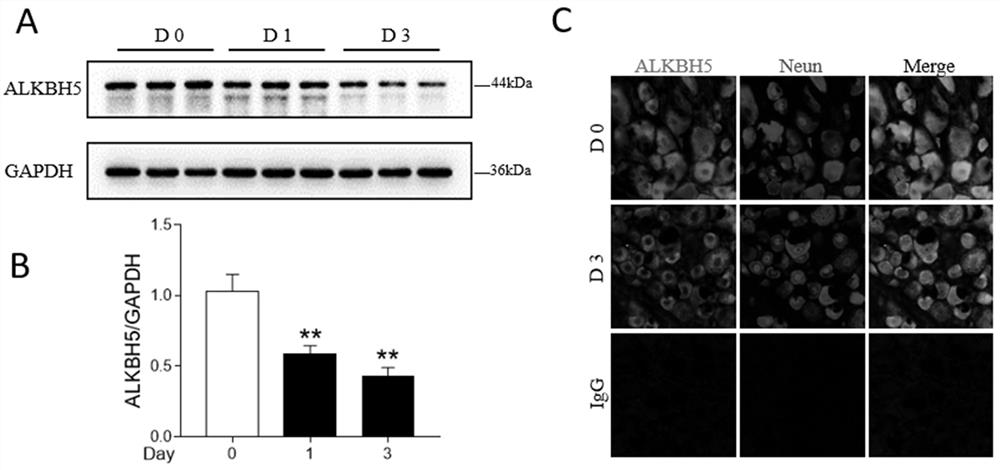
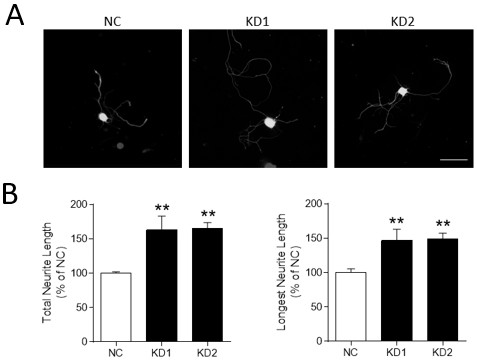
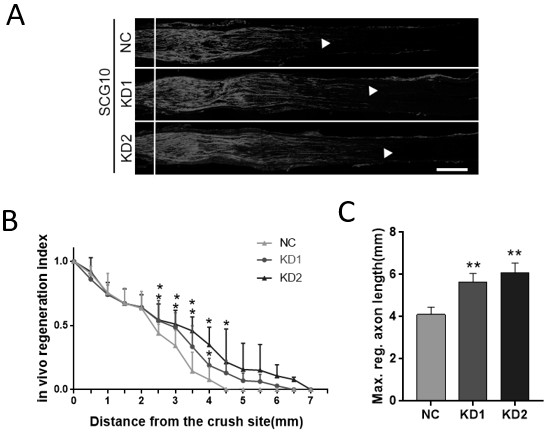
Application of m6a modification-related gene alkbh5 in promoting nerve axon repair
The invention discloses an application of m6A modification-related gene ALKBH5 in the preparation of medicines for treating diseases related to nerve axon damage. The invention also discloses an experimental verification of the application of m6A modification-related gene ALKBH5 in promoting the repair of nerve axon damage, which is characterized in that it comprises the following steps: S1, detecting the expression and location of ALKBH5; S2, interfering with the expression of ALKBH5 on nerve The effect of the axon growth; S3, the effect of interfering with the expression of ALKBH5 on the axon growth of the sciatic nerve; S4, the process of screening and functional testing of the ALKBH5 target gene LPIN2. The present invention verifies that interfering with ALKBH5 gene both in vivo and in vitro can significantly promote the growth of neuron axon, and promote the recovery of sciatic nerve function; and through transcriptome analysis and target gene verification after ALKBH5 interference, the target gene LPIN2 regulated by ALKBH5 is found and verified in Role in regulation of neuronal axon outgrowth.
Owner:NANTONG UNIVERSITY
A method for increasing the yield of streptomyces secondary metabolites
ActiveCN107805640BIncrease productionMicroorganism based processesFermentationBiosynthetic genesSecondary metabolite
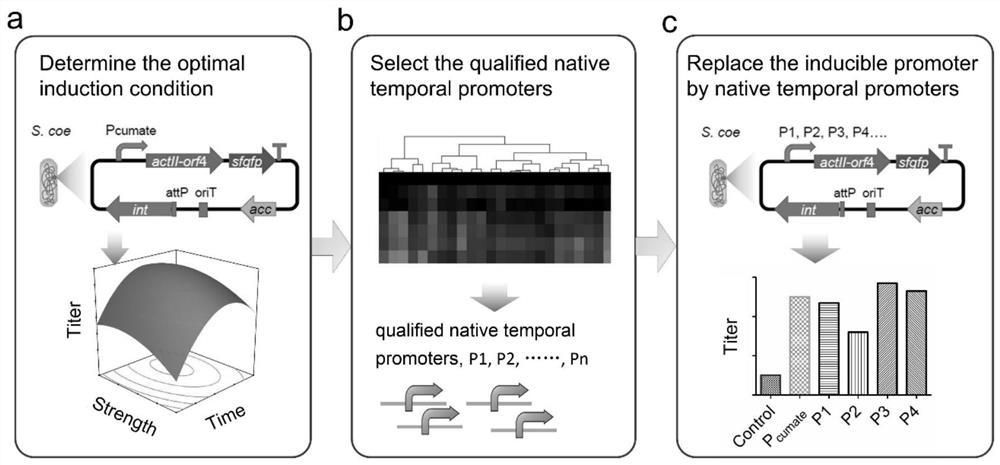
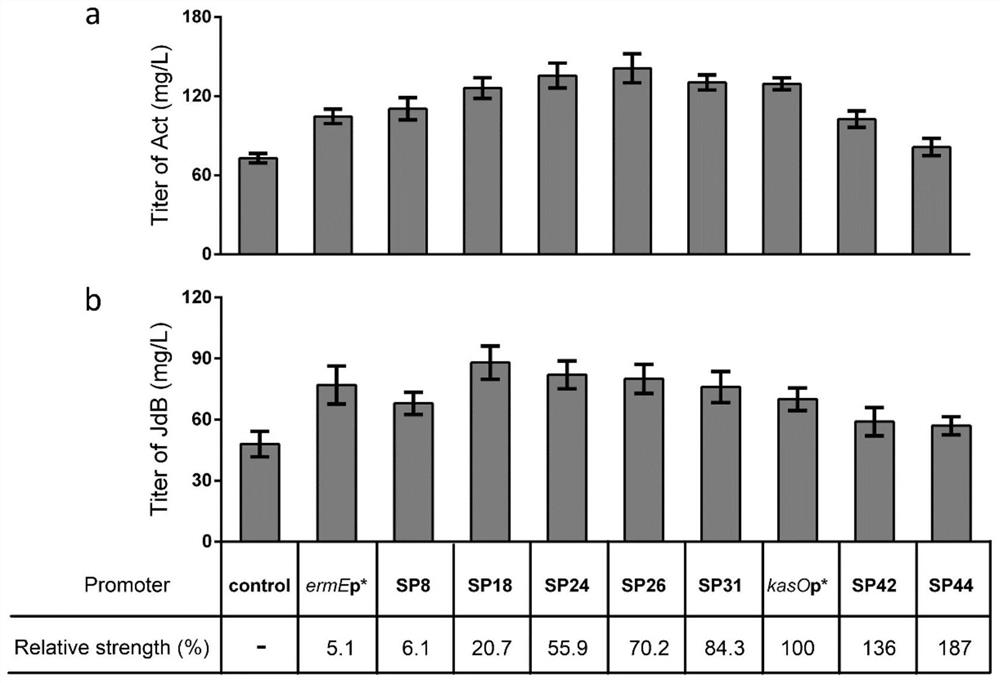
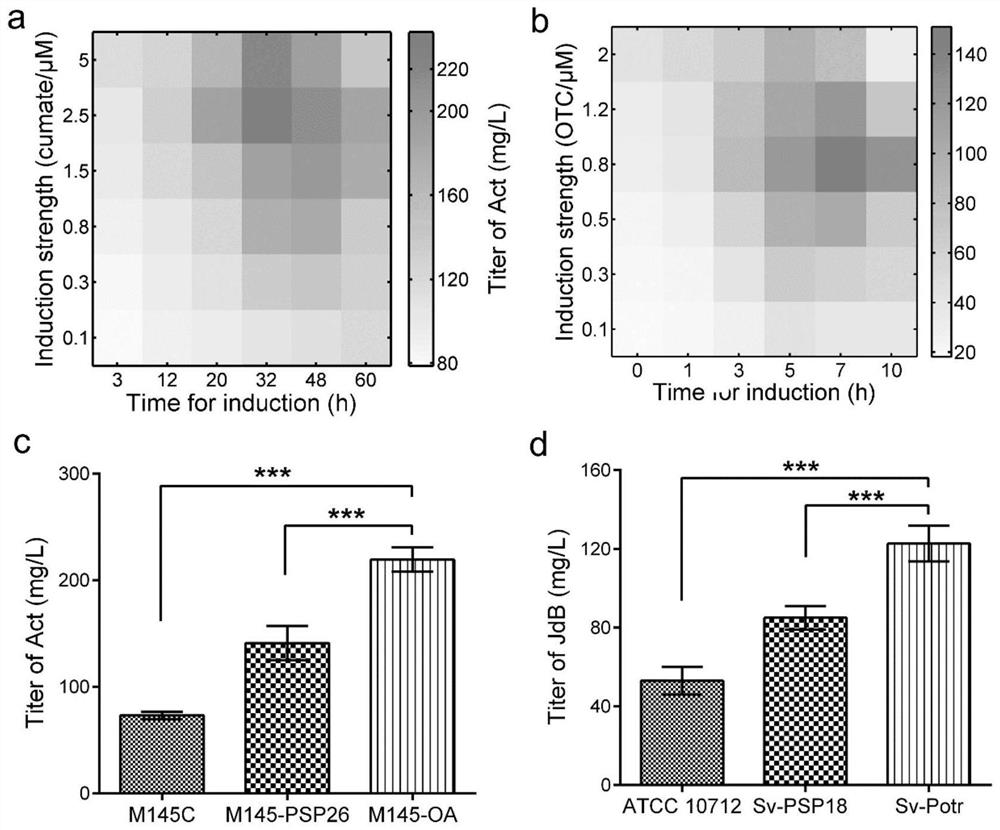
Belonging to the genetic engineering and fermentation engineering field, the invention provides a method for increasing streptomyces secondary metabolite yield. The method includes the steps of: utilizing an induction promoter to control the expression of a target secondary metabolite biosynthetic gene cluster, and determining the optimal induction condition by means of a response surface model; conducting transcriptome analysis to screen a physiological promoter with consistent control behavior to the induction promoter; and finally, preparing plasmid utilizing the physiological promoter to control the expression of the target secondary metabolite biosynthetic gene cluster, and transferring the plasmid into host bacteria for fermentation. The method provided by the invention can get rid of dependency on an inducer, and realizes self-regulation of target biosynthetic gene cluster expression and increase of the target product yield. The method provided by the invention regulates the expression of secondary metabolite biosynthetic gene cluster from the dimensions of time and intensity, makes the expression fit the physiological metabolic behaviors of the host, and is very important for increasing the yield of the target secondary metabolite in streptomyces.
Owner:INST OF PLANT PROTECTION CHINESE ACAD OF AGRI SCI
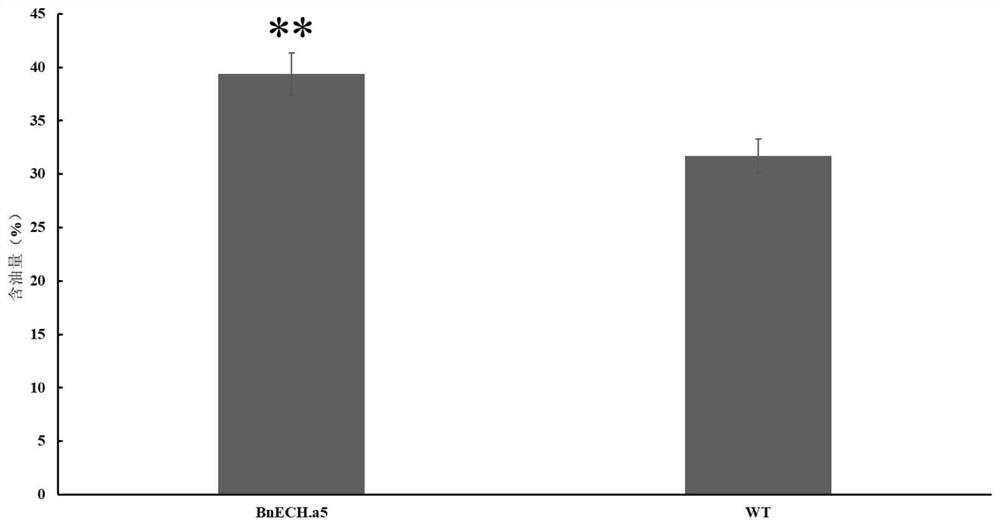
Method for rapidly identifying gene for regulating and controlling oil content of rape based on genomics approach and application thereof
PendingCN113667731AReduce workloadAccelerate cultivationMicrobiological testing/measurementPlant peptidesBiotechnologyGenetics genomics
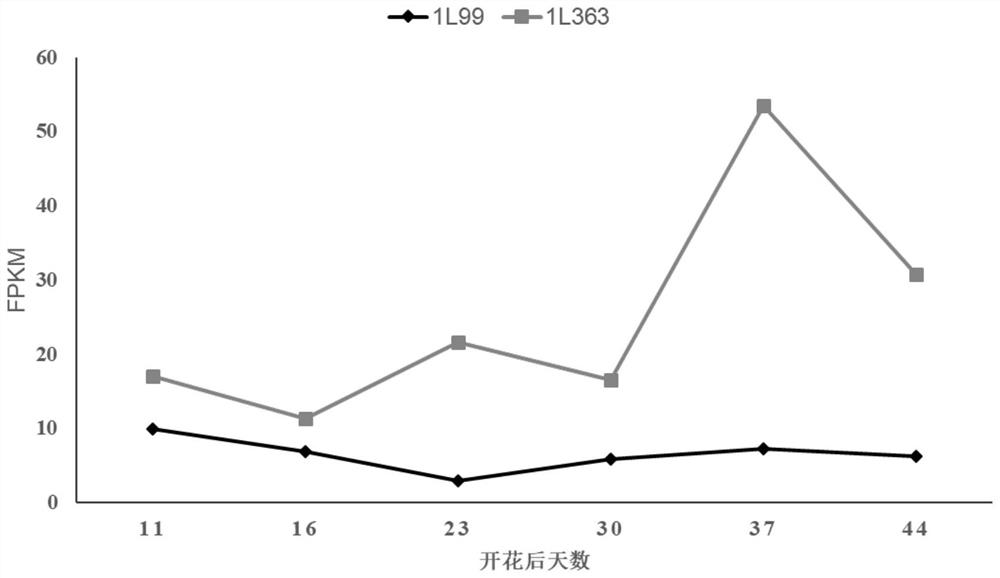
The invention belongs to the technical field of biology, and particularly relates to a method for rapidly identifying a gene for regulating and controlling the oil content of rape based on a genomics approach and an application of the method. According to the technical key points, the method comprises the following steps: screening a high-oil-content inbred line 1L99 and a low-oil-content inbred line 1L363 through field multi-year multi-point tests, and then carrying out transcriptome sequencing analysis on seeds of the two inbred lines in six different development stages; differential genes obtained through transcriptome analysis are scanned at a qOC-A5QTL site, and it is found that the expression quantity of the BnECH.A5 gene in different rape seed development stages of a high-oil-content inbred line is low, the expression quantity of the BnECH.A5 gene in a low-oil-content inbred line is high, and the expression quantity of the BnECH.A5 gene in the key stage of seed oil content accumulation within 30-44 days after flowering changes drastically. The mutant plant of the gene is constructed, the result shows that the oil content of a function-deleted mutant plant is remarkably increased compared with that of a wild plant.
Owner:HUAZHONG AGRI UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com