Screening and application of burkholderia gladioli real-time quantitative polymerase chain reaction (PCR) reference genes and primers thereof
A technology of Burkholderia Populus and internal reference gene, which is applied in the field of molecular biology, can solve the problems of deviation of experimental results and unstable RNA expression level of internal reference gene, and achieves the effects of high stability and wide adaptability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1B
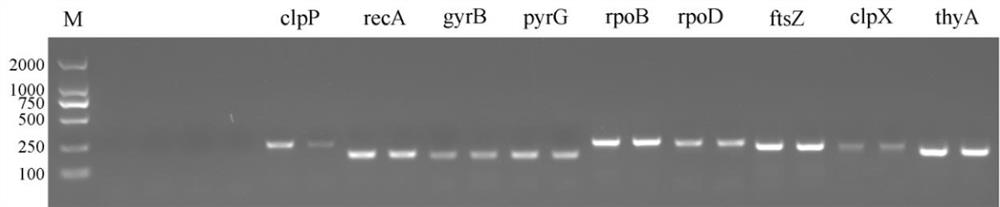
[0028] Screening of embodiment 1B.gladioli reference gene
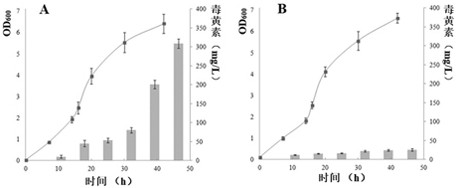
[0029] The strains used were B.gladioli HDXY-02 isolated and preserved in our laboratory and B.gladioli CICC10574 purchased from China Industrial Microbiology Culture Collection Center (CICC). The study found that B.gladioli HDXY-02 can produce obvious yellow pigment when using LB or KMB solid and liquid medium. After separation, extraction and identification, the pigment was determined to be toxin. Further research found that compared with the standard strain B.gladioli CICC 10574 purchased from CICC and B.gladioli HDXY-02, no yellow pigment was observed under either solid or liquid culture conditions. Growth curve and toxin synthesis analysis results (such as figure 1 shown), there is no significant difference in the growth of the two strains, but there is a significant difference in the toxin synthesis ability of the two strains (the bar graph is the toxin production), and B.gladioli HDXY-02 began to produce a la...
Embodiment 2
[0059] The expression stability analysis of the candidate internal reference gene of embodiment 2
[0060] 1. geNorm analysis
[0061] The geNorm software compares the genes in pairs to obtain the relative value of each gene change, and judges the stable value of the internal reference gene according to the M value. The lower the M value, the better the stability. Generally speaking, genes with an M value less than 1.5 indicate Its expression level is relatively stable.
[0062] Input the obtained Ct value into the geNorm program to obtain the expression stability value (M value) of each internal reference gene, and sort the internal reference genes according to the size of the M value. The results are shown in Table 2. At different culture stages, the pyrG and thyA genes of B.gladioli HDXY-02 were relatively stable, and the 16S and gyrB genes of B.gladioli CICC10574 were relatively stable; at different culture temperatures, the thyA genes of B.gladioli HDXY-02 and 16S genes...
Embodiment 3
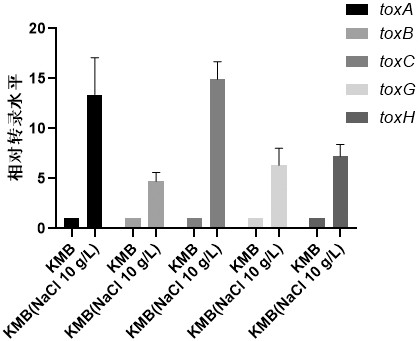
[0080] Example 3 Internal reference genes are applied to qRT-PCR to detect the expression of each gene in the tox gene cluster
[0081] The results of transcriptome sequencing (sampling point is the late logarithmic phase) showed that compared with B.gladioli CICC10574 as the control, the genes of the toxoflavin synthesis gene cluster toxABCDE and the transport gene cluster toxFGHI were significantly up-regulated in B.gladioli HDXY-02.
[0082] The transcriptome sequencing results were verified by qRT-PCR, and clpX was selected as an internal reference gene for qRT-PCR verification (see Table 6 for the primer sequences of genes related to toxoflavin synthesis). The experimental results were consistent with the results of transcriptome sequencing. Compared with B.gladioliCICC10574, the expression levels of toxoflavin synthesis genes toxA, toxB, toxC, toxD, toxE and transport genes toxF, toxG, toxH, toxI were lower in B.gladioli HDXY- 02 were increased by 5-10 times and 2-5 time...
PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com