Kit for detecting EB virus/HCMV and application thereof
An EB virus and kit technology, which is applied in the field of in vitro detection of organisms to achieve the effects of convenient clinical practice, wide detection range and simple operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
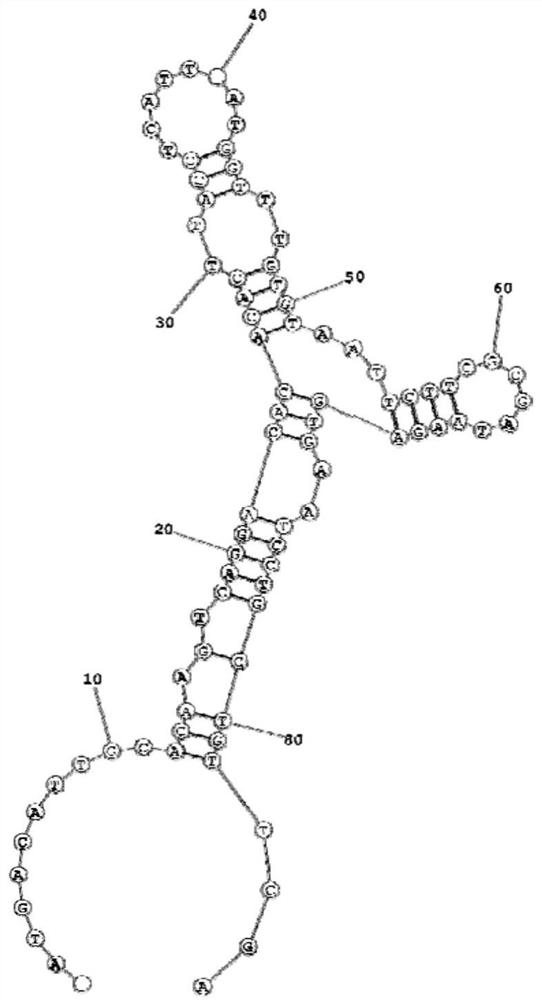
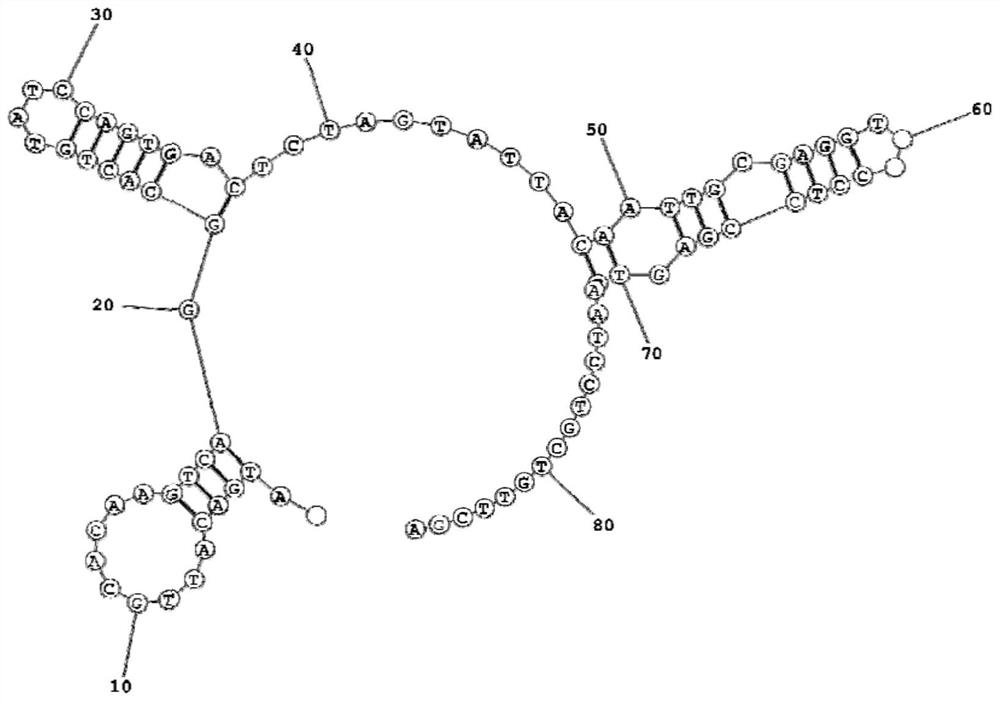
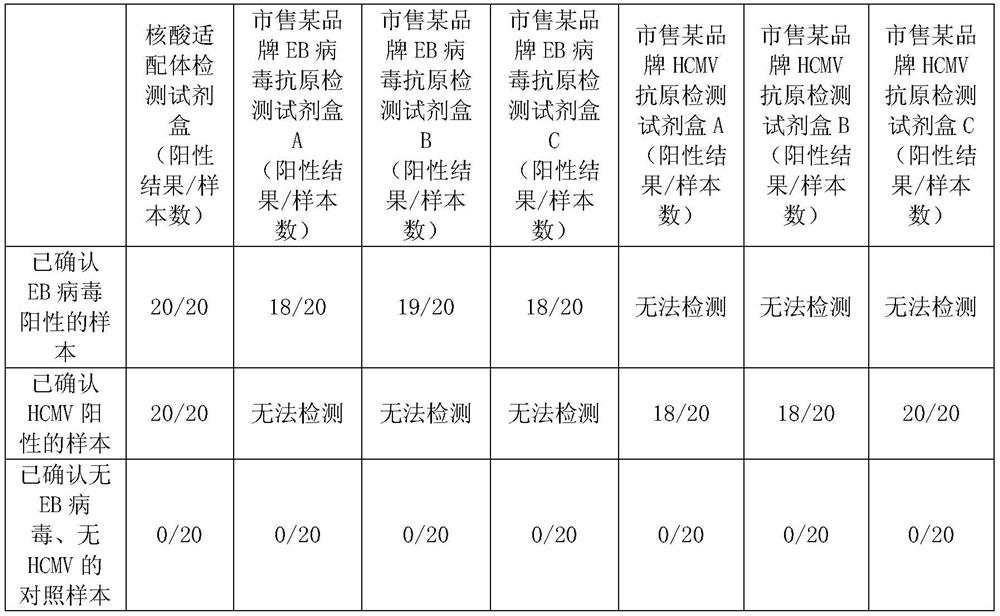
Image
Examples
Embodiment 1
[0037] Embodiment 1 determines the target protein epitope of Epstein-Barr virus, HCMV
[0038] According to literature review and bioinformatics analysis, the viral protein to be tested that is suitable as the detection target of the nucleic acid aptamer and the specific epitope region on the viral protein are determined.
[0039] A) Select the 400-857 region of the Epstein-Barr virus gp125 protein as an aptamer screening target protein, which has the sequence shown in SEQ ID No.3:
[0040] TTPTSSPPSSPSPPAPSAARGSTPAAVLRRRRRDAGNATTPVPPTAPGKSLGTLNNPATVQIQFAYDSLRRQINRMLGDLARAWCLEQKRQNMVLRELTKINPTTVMSSIYGKAVAAKRLGDVISVSQCVPVNQATVTLRKSMRVPGSETMCYSRPLVSFSFINDTKTYEGQLGTDNEIFLTKKMTEVCQATSQYYFQSGNEIHVYNDYHHFKTIELDGIATLQTFISLNTSLIENIDFASLELYSRDEQRASNVFDLEGIFREYNFQAQNIAGLRKDLDNAVSNGRNQFVDGLGELMDSLGSVGQSITNLVSTVGGLFSSLVSGFISFFKNPFGGMLILVLVAGVVILVISLTRRTRQMSQQPVQMLYPGIDELAQQHASGEGPGINPISKTELQAIMLALHEQNQEQKRAAQRAAGPSVASRALQAARDRFPGLRRRRYHDPETAAALLGEAETEF(SEQ ID No.3)
[0041] B) Select the ...
Embodiment 2
[0044] Example 2 Construction of aptamer library and primer design
[0045] A) A single-stranded DNA library containing 45 random sequences was synthesized in vitro through the gene synthesis service of a conventional biotechnology service company, and its nucleotide sequence is as follows
[0046] GATGACATTGCACAAGTCAGG-(N45)-GAGTGAATCCTGCTGTTCGA
[0047] B) For the fixed sequence at the 5' end of the nucleic acid adapter, design a synthetic upstream primer P1, which has a sequence as shown in SEQ ID No.5:
[0048] 5'-GATGACATTGCACAAGTCAGG-3' (SEQ ID No.5)
[0049] C) Aiming at the fixed sequence at the 3' end of the nucleic acid aptamer, design a synthetic downstream primer P2, which has the sequence shown in SEQ ID No.6, and a biotin group is attached to the 5' end of the primer:
[0050] 5'-biotin-TCGAACAGCAGGATTCACTC-3'(SEQ ID No.6)
Embodiment 3
[0051] Example 3 Using SELEX screening technology to screen the library for nucleic acid aptamer sequences that can effectively bind to viral target proteins
[0052] A) Dissolve 10 nmol of the initial single-stranded DNA random library in 500 μl of PBS solution, bathe in a constant temperature water bath at 92°C for 5 minutes, then quickly insert it into ice for 10 minutes, and then place the treated initial single-stranded DNA random library Incubate with the corresponding virus target protein on ice for 1h, then add Ni-NTA Magnetic Agarose Beads, and continue to incubate for 1h. After incubation, use a magnetic separator to absorb, remove the supernatant, and wash the Beads with 2ml of PBS. Finally, resuspend the Beads with 10ml of pre-cooled PBS to make them fully blown and mixed, and then in a constant temperature water bath at 92°C for 10 minutes, centrifuge at 13,000g, and collect the supernatant, which is the single-stranded DNA library that specifically recognizes the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com