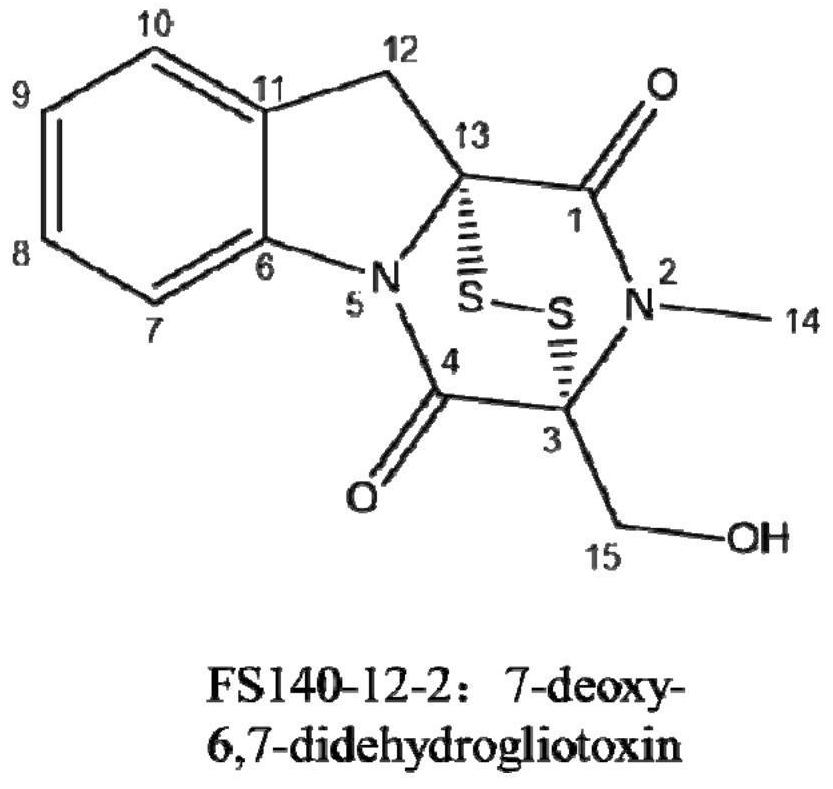
Deep-sea fungus FS140 anti-gliotoxin self-protection gene GliM and application thereof
A gliotoxin and gene protection technology, applied in the field of genetic engineering, can solve the problems of inducing apoptosis, redox reaction imbalance, reducing immune activity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1 Obtaining the self-protection gene sequence of the deep-sea fungus Geosmithia pallida FS140 against gliotoxin
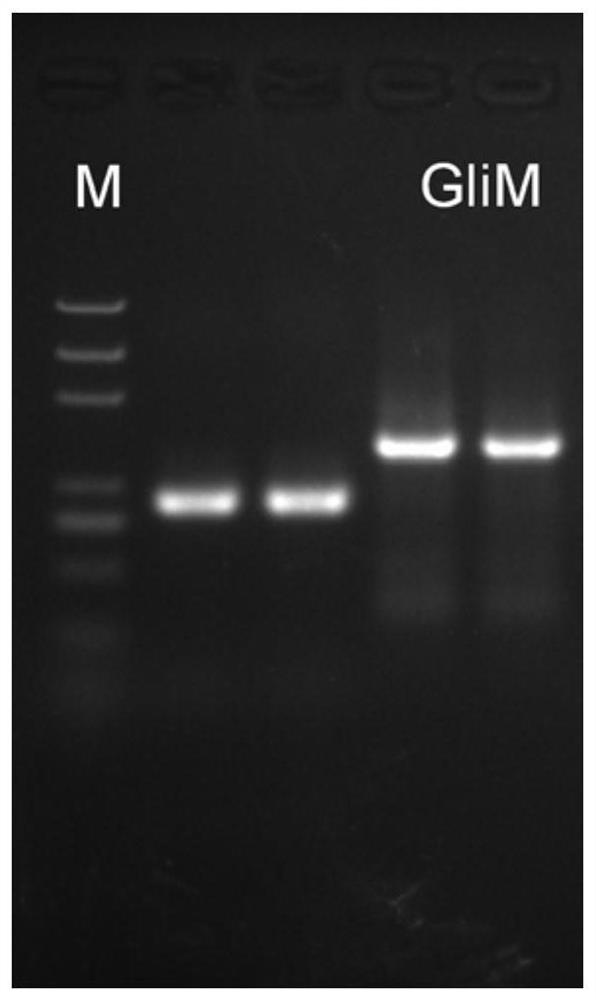
[0025]Amplification of the gene GliM: Inoculate the deep-sea fungus Geosmithia pallida FS140 on the YPD medium plate, culture at 37°C for 72 hours, pick fresh mycelium, use the fungal RNA extraction kit to extract RNA, and then use the All-in-one RTMaster cDNA was obtained by reverse transcription with Kit. According to the transcriptome sequencing results, the anti-trichothecene self-protection gene GliM sequence was predicted, and specific primers were designed upstream and downstream of it. The primer sequence was GliM-F:5'-ATGGAAGCCAACAACACCGAC-3'; GliM-R:5' -CTACTTCTTCAGCCGTAATTCCAA-3', using the cDNA library as template amplification to obtain PCR product ( figure 1 ). The product was recovered and cloned by TA with pEASY-T1 kit, transformed into Escherichia coli competent cells, spread on the ampicillin resistance plate to screen out positi...
Embodiment 2
[0026] Example 2 Functional verification of anti-gliotoxin self-protection gene GliM
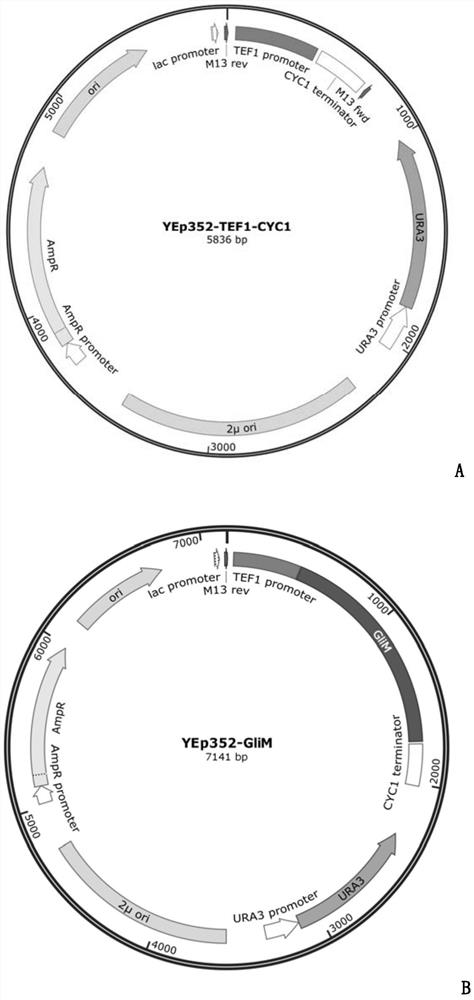
[0027] The gene GliM was inserted into the yeast vector YEp352-TEF1-CYC1 by homologous recombination (YEp352-TEF1-CYC1 is an early constructed plasmid, carrying a constitutive promoter TEF1 and a terminator CYC1, see the vector map image 3 A, known products in the prior art: Xiaodan Ouyang, Yaping Cha, Wen Li, Chaoyi Zhu, Muzi Zhu, ShuangLi, Min Zhuo, Shaobin Huang and Jianjun Li. Stepwise engineering of Saccharomyces cerevisiae to produce(+)-valencene and its related sequiterpenes, RSC Adv., 2019, 9, 30171, DOI: 10.1039 / c9ra05558d). First design the upstream and downstream primers YEp352-GliM-F and YEp352-GliM-R for the amplification of gene GliM (SEQ ID NO.1), the primer sequence of which is YEp352-GliM-F:5'- ATAGCAATCTAATCTAAGTCTAGA ATGGAAGCCAACAACACCGAC-3'; YEp352-GliM-R:5'- TACATGATGCGGCCCGTCGAC CTACTTCTTCAGCCGTAATTCCAA-3' (the underlined sequence is the homology arm fragment), the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com