Proteome obtained by splitting Cas9 protein and application of proteome
A proteomics and protein technology, applied in the field of proteomics, can solve the problems of reduced splicing efficiency and no longer applicable, and achieve the effects of improving gene editing efficiency and safety, wide selection range, and low off-target rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0087]Example 1 Split of Cas9 protein
[0088]The CAS9 protein was split into two different amino acid sequences CAS9N (N end) and CAS9C (E-terminal). Among them, the CAS9 protein is SPCas9 (H840A) protein, and its amino acid sequence is shown in SEQ ID NO.1, and the encoding nucleotide sequence is shown in SEQ IDNO.2.
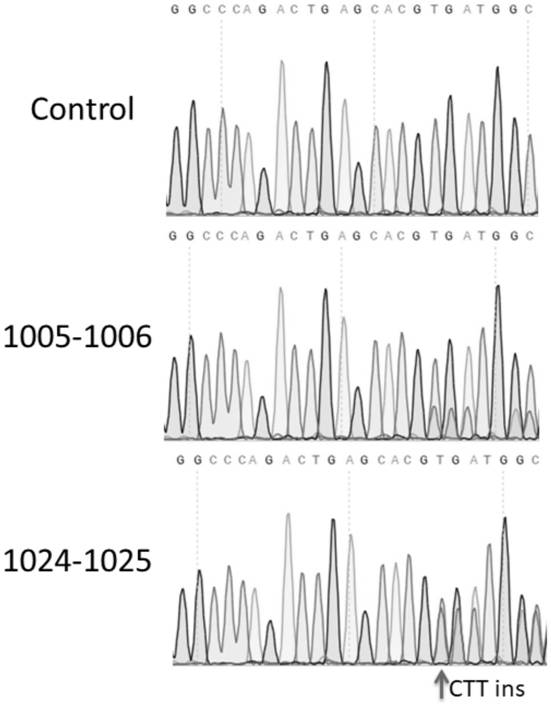
[0089]The split positions are: between the 994th to 995 bits, between the 1,05 to 1006 bits, between the 1024th to 1025 bits, and between the 1032 to 1033 bits, the unino acid sequences obtained by each of the detachment fragments obtained and The nucleotide sequence encoding it is shown in Table 1.
[0090]Table 1:
[0091]
Embodiment 2
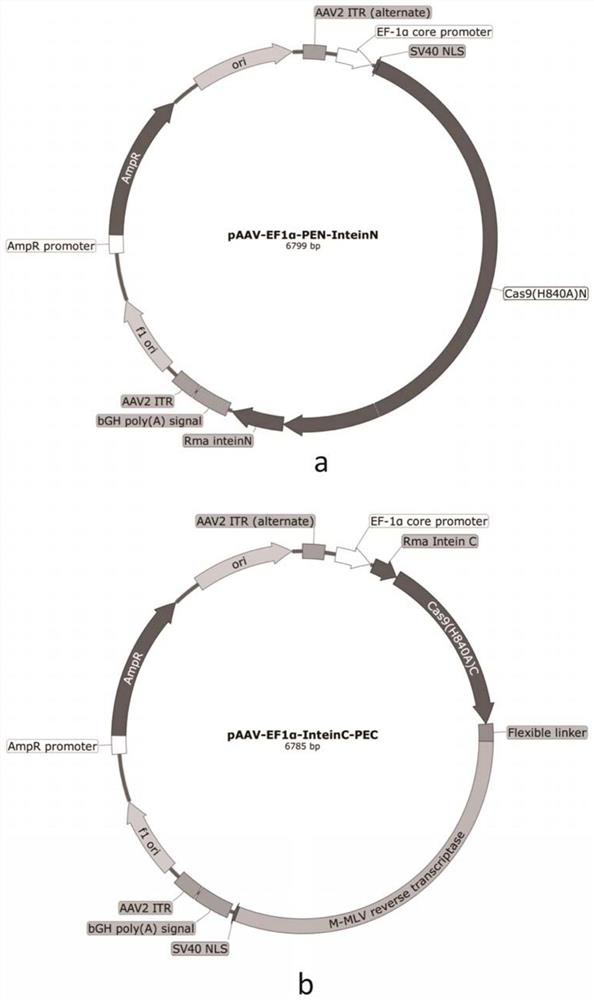
[0092]Example 2 Construction of AAV carrier
[0093]First, experimental method
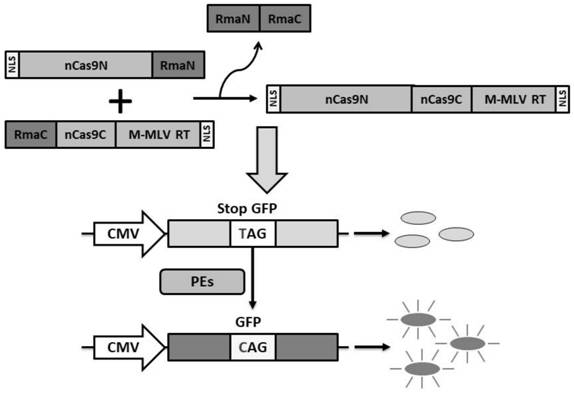
[0094]1. Split the peptide RMA INTEIN gene (whose nucleotide sequence such as SEQ ID NO.7 is shown, which is shown in SEQ ID No. 6, as shown in SEQ ID NO. The RMAC, the sequence is the nucleotide sequence fragment of the first to 306th position of the nucleotide sequence shown in SEQ IDNO 7, respectively, and the nucleotide sequence of the No. 307 to 462 (corresponding amino acid fragment, respectively, respectively, respectively, respectively.) SEQ ID NO. 6 The amino acid fragment of 1 to 102 of the amino acid sequence shown and the amino acid fragment of No. 103 to 154) of the amino acid sequences.
[0095]2. According to the Cas9 split method of Table 1, Prime Editor (PE) is constructed, and the nucleotide sequence is shown in SEQ IDNO.10, wherein the sequence of the first to 27th base is a nucleoside encoding a nuclear positioning signal peptide. The sequence of acid sequences (encoded amino terminal sequenc...
Embodiment 3
[0108]Example 3 Splitted PE system can effectively perform editing of a karace point point
[0109]First, experimental method
[0110]The split-PE system was constructed in the method of Example 2 containing the expression vector group containing reverse transcriptase: PAAV-EF1α-Pen-INTEINN and PAAV-EF1α-INTEINC-PEC, wherein the expression carrier group is SPCas9 protein, respectively, according to respectively. Split from the following four sites: Between the 994th to 995 bits, between the 1,05-006 bits, between the 1024th to 1025 bits or between the 1st 332 to 1033, the peptide is RMA INTEIN.
[0111]HEK293T cells containing M1EMGFP steady expressed, M1EMGFP for EMGFP (Nucleotide Sequence, SEQ ID NO.11) Sequence Codon CAG mutant to TAG, due to the EMGFP mutation, causes the translation to terminate in advance, only Tag is edited to CAG to restore the normal expression of GFP, so the ability of gene editing can be detected by statistics of GFP positive cells and accurate and convenientness ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com