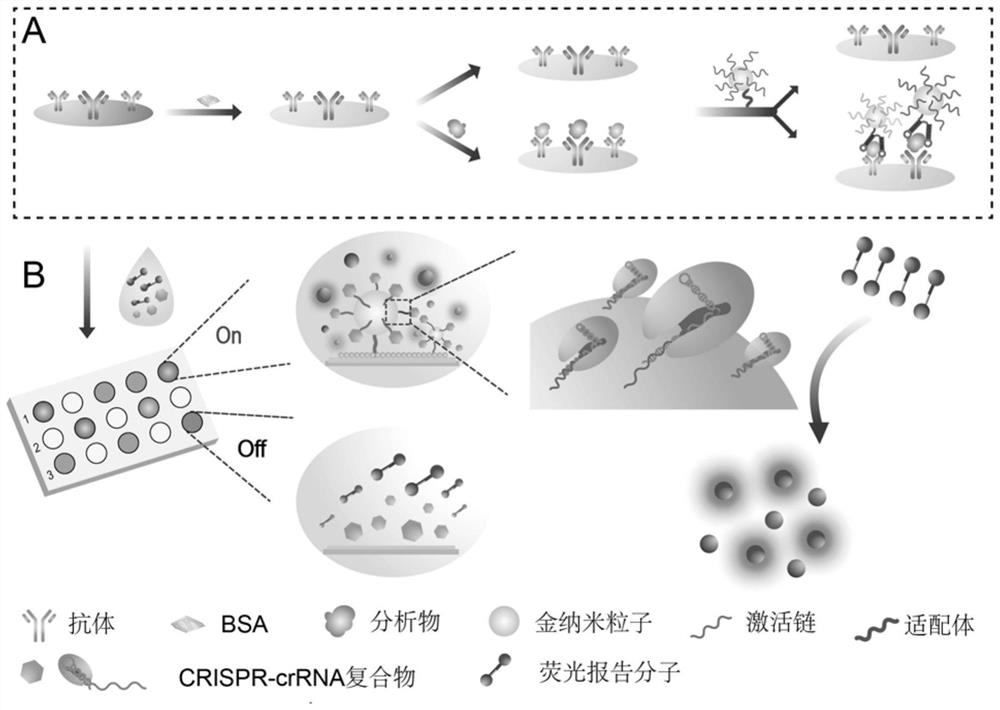
Protein marker detection kit combining gold nanoprobe and CRISPR-Cas and detection method
A protein marker and gold nanotechnology, which is applied in the chemical and biological fields, can solve the problems of sample cross-contamination, complicated operation procedures, and increased reaction costs, and achieve the effects of preventing cross-contamination, simplifying the operation process, and widening the linear range of detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] In this example, CEA was used as an analyte for quantitative analysis.
[0050] 1. Preparation of gold nanoprobes
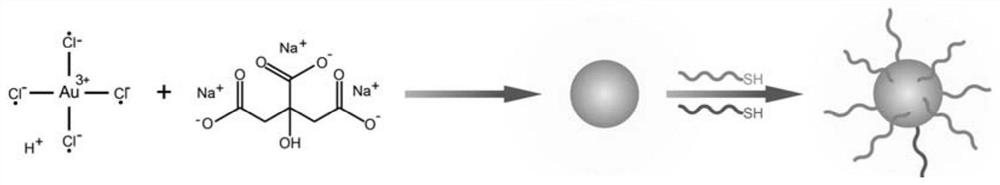
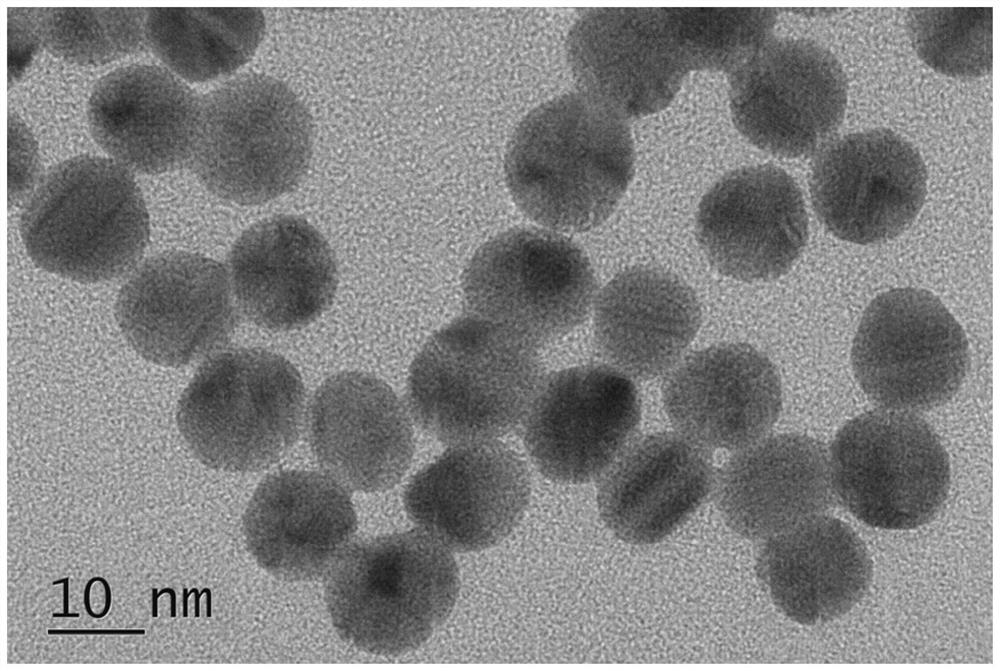
[0051] The preparation process of gold nanoprobes is as follows: figure 2 As shown, the CEA aptamer and the activated chain were activated with 10 mM TECP for 1 hour at room temperature, and then they were added to the gold nanoparticle solution and reacted with shaking at room temperature for 16 hours. Then add 1M NaCl solution six times to make the final concentration 0.1M, shake at room temperature for no less than 24 hours to obtain gold nanoprobes, centrifuge at 10000r / min for 10min to remove excess nucleic acid, and use PBS (0.01M, pH 7.4) buffer solution was washed three times, redispersed in PBS (0.01M, pH 7.4) buffer solution, and stored at 4°C in the dark for future use. like image 3 UV characterization, indicating the successful preparation of gold nanoprobes.
[0052] The sequence of the CEA aptamer is:
[0053] 5'-SH-C 6 -TTTTTTTTTTTAA...
Embodiment 2
[0072] In this example, PSA was used as an analyte for quantitative analysis.
[0073] The difference from Example 1 is that the CEA aptamer is replaced with the PSA aptamer, and the specific coating antibody of CEA is replaced with the specific coating antibody of PSA.
[0074] The sequence of the PSA aptamer is:
[0075] 5'-SH-C 6 -TTTTTAATTAAAGCTCGCCATCAAATAGC-3'
[0076] The PSA was purchased from Shanghai Lingchao Biotechnology Co., Ltd., diluted with PBS (0.01M, pH 7.4) buffer solution to different concentrations: 0, 0.01, 0.05, 0.1, 0.5, 1, 5, 10, 30, 50, 70, 100, 150ng / mL.
[0077] Figure 6 The standard curve of PSA is shown, and it can be seen from the figure that the signal of the detection system increases with the increase of PSA analyte, and there is a good linear relationship between 0.5-150 ng / mL.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com