SNP Molecular Markers Related to Maize Row Kernel Number and Its Application
A molecular marker, row number technology, applied in recombinant DNA technology, microbial determination/inspection, DNA/RNA fragments, etc. process, the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026]1.1 Construction of GWAS groups
[0027] The maize genome-wide association analysis population includes 431 inbred lines with different kinship. All inbred lines are provided by the School of Plant Science, Jilin University, and are planted in the Teaching and Research Experimental Base of the School of Plant Science, Jilin University (Lvyuan District, Changchun City, Jilin Province). The arrangement of the plots followed a random block design, the block was repeated 3 times, the single-row plot, the row length was 3 m, the row spacing was 65 cm, and the plant spacing was 20 cm. The field management measures were the same as those in the field.
[0028] 1.2 Collection of samples and investigation of phenotypic data
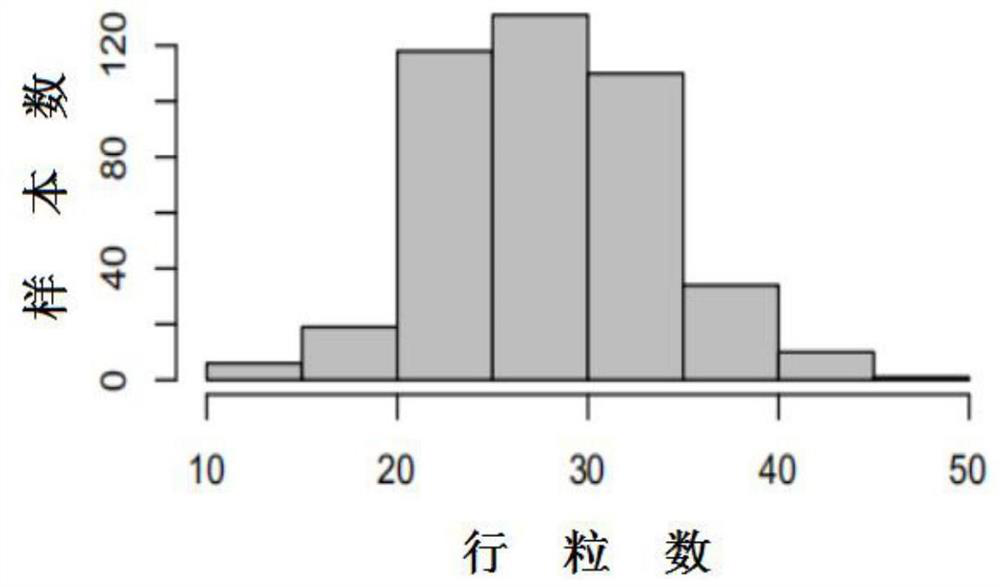
[0029] At the 5-6 leaf stage of the maize seedlings in the plot, 3 seedlings were collected, cleaned and stored in a -20°C refrigerator. Five representative ears were harvested from each plot, and the number of kernels per row (KPR) was examined after the ...
Embodiment 2
[0041] 2.1 Construction of sequencing library
[0042] (1) 200ng of genomic DNA was digested with the restriction enzyme EcoRI, and after the digestion was complete, it was purified and recovered by magnetic beads.
[0043] (2) The purified DNA after enzyme digestion was connected to Barcode adapters PI with T4 DNA Ligase, the ligation product was purified and recovered by magnetic beads, and the qualified P1 primer label was recorded.
[0044] (3) All samples were mixed in equal proportions into a total DNA mix, and the DNA was fragmented using covaris220. The fragment length of the fragmented DNA was about 200-500 bp.
[0045] (4) After End Repair & dA-tailing, connect to Adaptor P2, and purify and recover the ligated product with magnetic beads.
[0046] (5) Use the KAPA 2G Robust PCR Kit to amplify and enrich the linker ligation products, use magnetic beads to purify and sort the PCR reaction products, and use 2% agarose gel electrophoresis to detect the quality of the co...
Embodiment 3
[0063] Intergroup comparison of different genotypes detected at significant loci
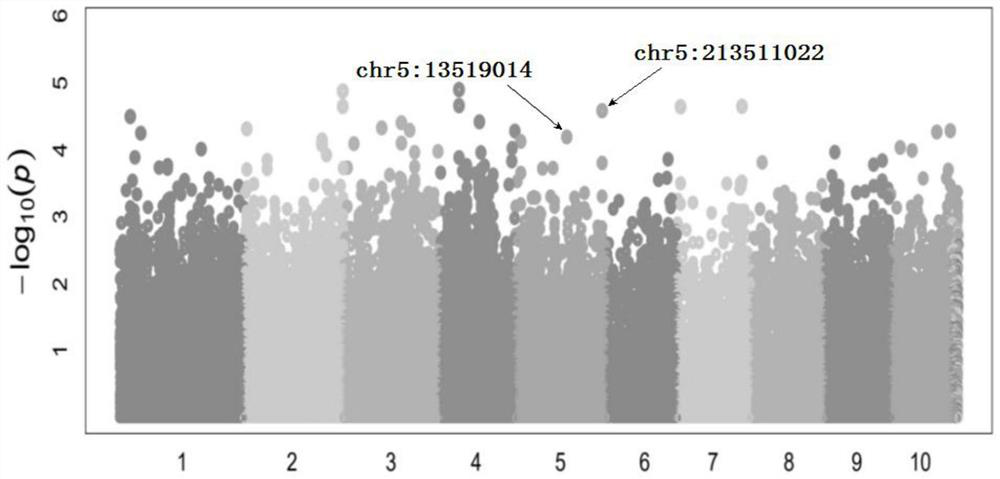
[0064] 3.1 Zm00001d018060 gene Chr5:213511022 locus comparison between groups
[0065] This locus includes three genotypes, namely C / C, T / T, and C / T. The box plots of the row grain number distribution of samples of different genotypes are shown in Figure 5 . The median of C / C group was 25.77 grains, the lower quartile Q1 was 22.83 grains, and the upper quartile Q3 was 29.77 grains; the T / T group had a median of 28.66 grains, and the lower quartile Q1 was 24.77 grains grains, the upper quartile Q3 was 31.00 grains; the median of C / T group was 28.55 grains, the lower quartile Q1 was 25.00 grains, and the upper quartile Q3 was 32.05 grains.
[0066] The t-test results of the phenotypic mean difference between different genotype groups are shown in Table 2. It can be seen from Table 2 that the difference between the C / C and T / T groups is extremely significant (P=0.0001), and the mean difference b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com