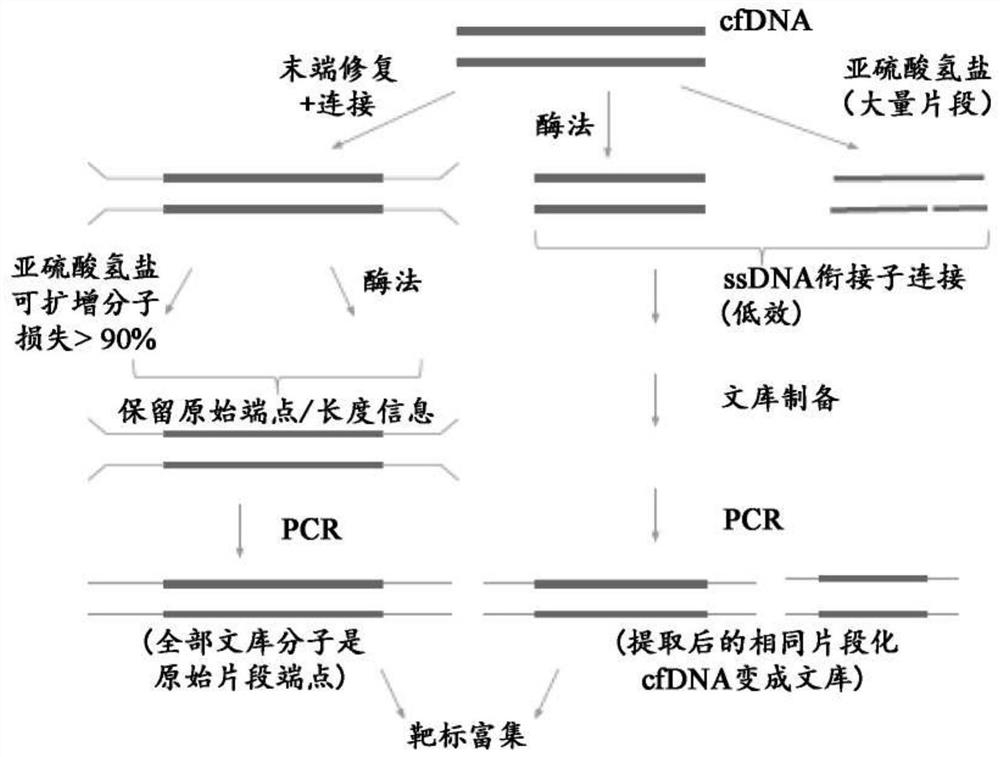
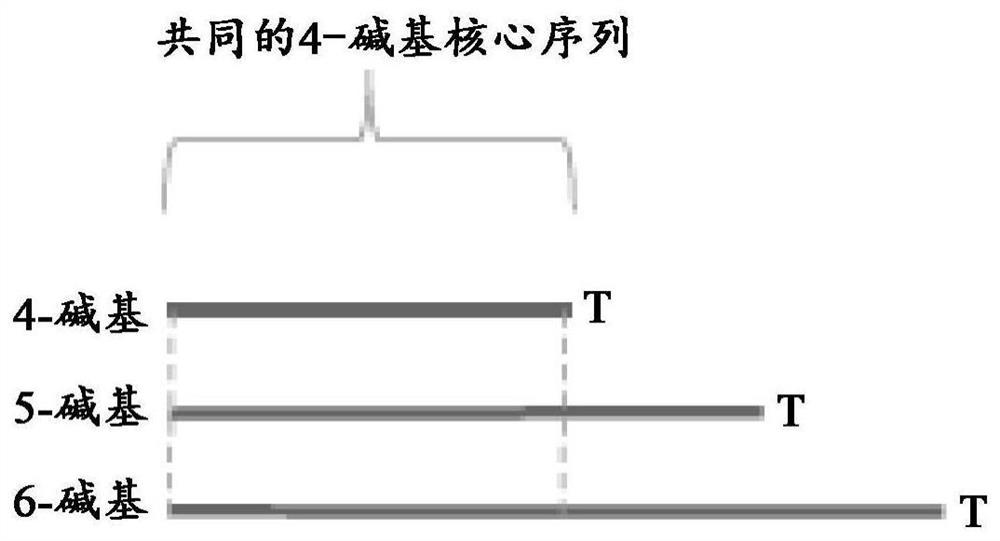
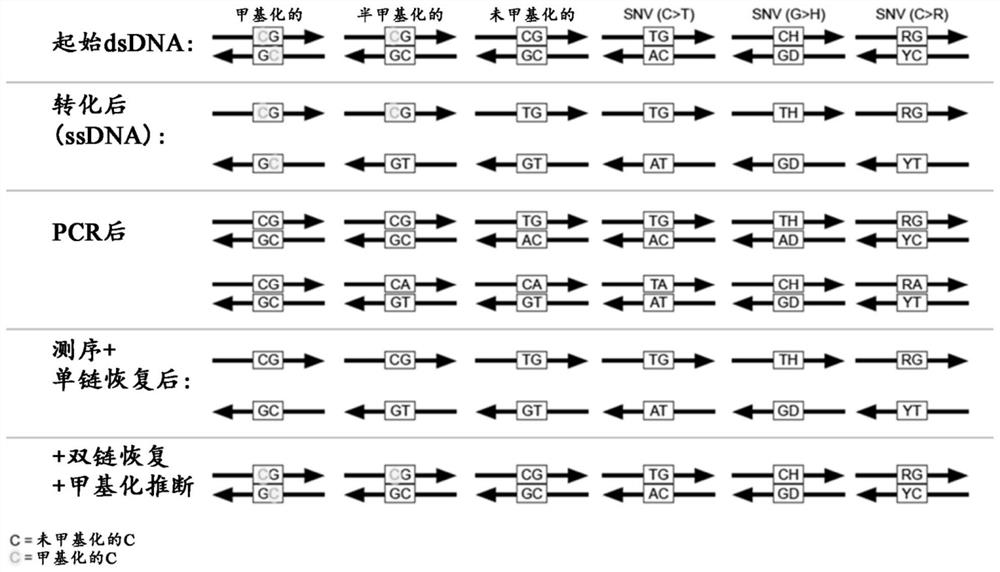
Methods and systems for high-depth sequencing of methylated nucleic acid
A methylated and unmethylated technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0377] Example 1: Targeted EM sequencing library preparation and classifier generation
[0378] Starting material: 10-200 ng of double-stranded DNA.
[0379] 1. DNA preparation
[0380] EDTA was removed from the DNA before oxidation and the DNA sample had a final volume of 29 μl. Control DNA was used to assess oxidation and deamination. For sequencing on the Illumina platform, refer to the enzymatic methyl sequencing kit manual (NEB#E7120) for usage recommendations.
[0381] 2. Adapter ligation
[0382] 3. Oxidation of 5-methylcytosine and 5-hydroxymethylcytosine
[0383] TET2 buffer was prepared. TET2 Reagent was then added to a tube with TET2 Reaction Buffer Supplement followed by thorough mixing. On ice, add TET2 Reaction Buffer, Oxidation Supplement, Oxidation Promoter, and TET2 Enzyme directly to the DNA sample. The mixture was then mixed well by vortexing. After a brief centrifugation, the diluted Fe(II) solution was added to the mixture. The mixture was then mi...
Embodiment 2
[0395] Example 2: Targeted EM Sequencing Using a Transformation-Resistant Sequencing Adapter / Primer System
[0396] Identified adapters of known sequence are ligated to the ends of DNA molecules containing unknown sequences in the sample. The adapters are then used to PCR amplify the entire library of different molecules using a single primer set corresponding to the known adapters. The ligated adapter sequences also serve as binding sites for sequencing primers during subsequent sequencing reactions. To utilize the data provided by double-stranded sequencing, partially double-stranded adapters containing unique molecular identifiers (UMIs) were ligated to double-stranded DNA.
[0397] To improve the robustness, oxidation efficiency (and reduce cost) of EM-sequencing double-stranded sequencing, transformation-resistant adapters can be used to increase the consistency of sequencing library quality. Conversion-resistant adapters contain only unmodified bases and allow complete...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com