Construction method, kit and application of plasma free DNA methylation detection library
A construction method and methylation technology, which is applied in the field of plasma cell-free DNA methylation detection library construction, can solve the problems that plasma cell-free DNA cannot construct a library, reduce the initial amount of library construction, improve library recovery efficiency, simplify The effect of the library building step
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0028] According to a typical embodiment of the present invention, a method for constructing a plasma cell-free DNA methylation detection library is provided. The construction method includes the following steps: S1, extracting plasma free DNA from blood samples; S2, performing end repair on plasma free DNA and adding A base to the 3' end; S3, connecting the end of plasma free DNA obtained in S2 to C Modified linker (design principle: phosphorylation at the 5' end of the F chain, all C methylation modifications, all C methylation modifications on the R chain); S4, perform bisulfite conversion on the product obtained from S3, and perform PCR extension; S5, perform magnetic bead purification on the PCR product of S4, remove small fragments of DNA and primer dimers that have not been non-specifically amplified; and S6, perform PCR amplification on the product of S5, and perform magnetic bead purification, Obtain plasma cell-free DNA methylation detection library.
[0029] Prefer...
Embodiment 1
[0045] The main experimental steps are as follows:
[0046] 1. Acquisition of plasma samples
[0047] Under normal laboratory conditions, use EDTA blood collection tubes, preferably Streck blood collection tubes to collect 10ml of peripheral blood from prostate cancer patients, centrifuge quickly at room temperature for 10min at 1900g (or 3000rpm), and gently draw the upper layer of plasma (to avoid buffy coat contamination) into a new 15ml centrifuge tube , centrifuge at 16000g room temperature for 10min, and gently pipette the supernatant into a new 15ml centrifuge tube.
[0048] 2. Extraction and quality inspection of plasma cell-free DNA
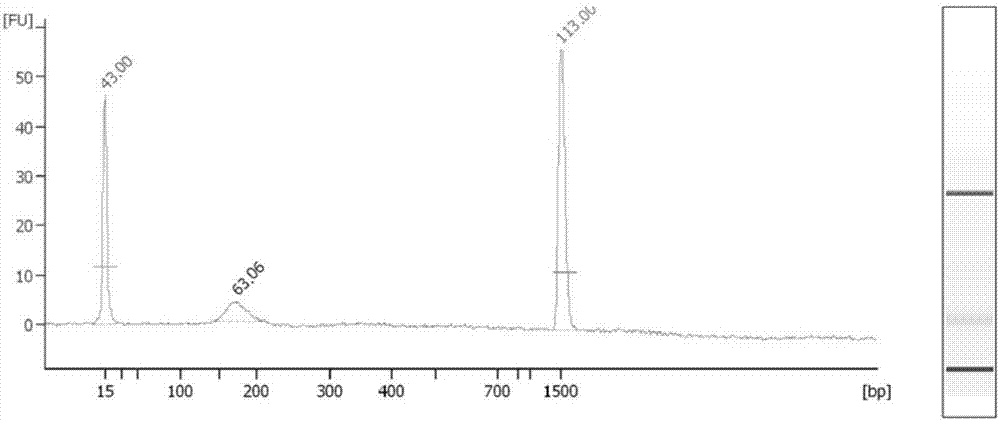
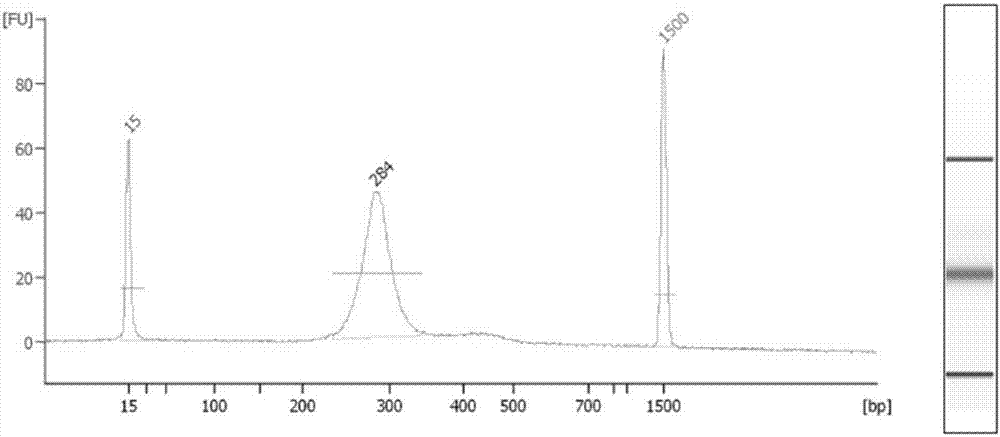
[0049] Use Qiagen’s kit for extracting free nucleic acid under routine laboratory conditions, preferably Circulating Nucleic Acid kit, extract free DNA from the separated plasma according to the instructions, and elute with 30 μl warm water, and use Agilent 2100 to qualitatively and quantitatively detect the extracted plasma free DNA,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com