A method for simultaneously hydrolyzing and crosslinking self-porous polymer membranes
A polymer and polymer material technology, applied in the field of polymer science, can solve the problems of long cross-linking time (thermal cross-linking generally takes at least 3 days, loss of membrane mechanical properties, reduction of membrane permeability coefficient, etc.), and achieve excellent gas separation. Ability, mild conditions, novel effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
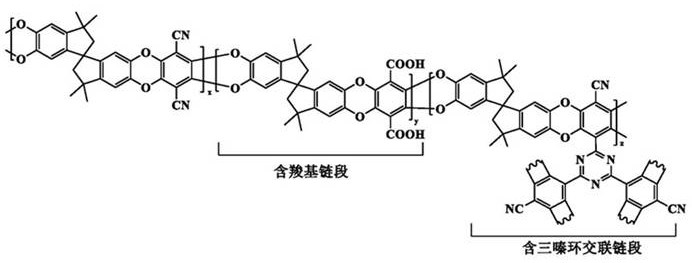
[0029] Soak the PIM-1 membrane prepared in the second step of Comparative Example 1 in methanesulfonic acid for 10 min, then place the membrane soaked in methanesulfonic acid on a glass plate, and then transfer the glass plate to an oven at 110°C for PIM -1 The hydrolysis and crosslinking reaction of the film, the reaction time is 12h. After the reaction was completed, the membrane was taken out from the oven, soaked and rinsed in deionized water, and dried to obtain a novel membrane cPIM-1 containing carboxyl and triazine groups. In the cPIM-1 membrane structure prepared in Example 1, 8% of the nitrile groups were cross-linked to form a triazine ring, and the degree of cross-linking of the membrane was 8%.
[0030] The obtained cPIM-1 structural formula is shown in the figure below:
[0031]
[0032] The second step, the gas separation performance characterization of cPIM-1: test the cPIM-1 membrane to N 2 、CH 4 , CO 2 Gas separation performance.
[0033] See Table 1 ...
Embodiment 2
[0035] Soak the PIM-1 membrane prepared in the second step of Comparative Example 1 in methanesulfonic acid for 10 min, then place the membrane soaked in methanesulfonic acid on a glass plate, and then transfer the glass plate to an oven at 110°C for PIM -1 membrane hydrolysis and crosslinking reaction, the reaction time is 24h. After the reaction was completed, the membrane was taken out from the oven, soaked and rinsed in deionized water, and dried to obtain a novel membrane cPIM-1 containing carboxyl and triazine groups. In the cPIM-1 membrane structure prepared in Example 2, 28% of the nitrile groups were crosslinked to form a triazine ring, and the degree of crosslinking of the membrane was 28%.
[0036] The second step, the gas separation performance characterization of cPIM-1: test the cPIM-1 membrane to N 2 、CH 4 , CO 2 Gas separation performance.
[0037] See Table 1 for the gas separation performance of the cPIM-1 membrane prepared in Example 2.
Embodiment 3
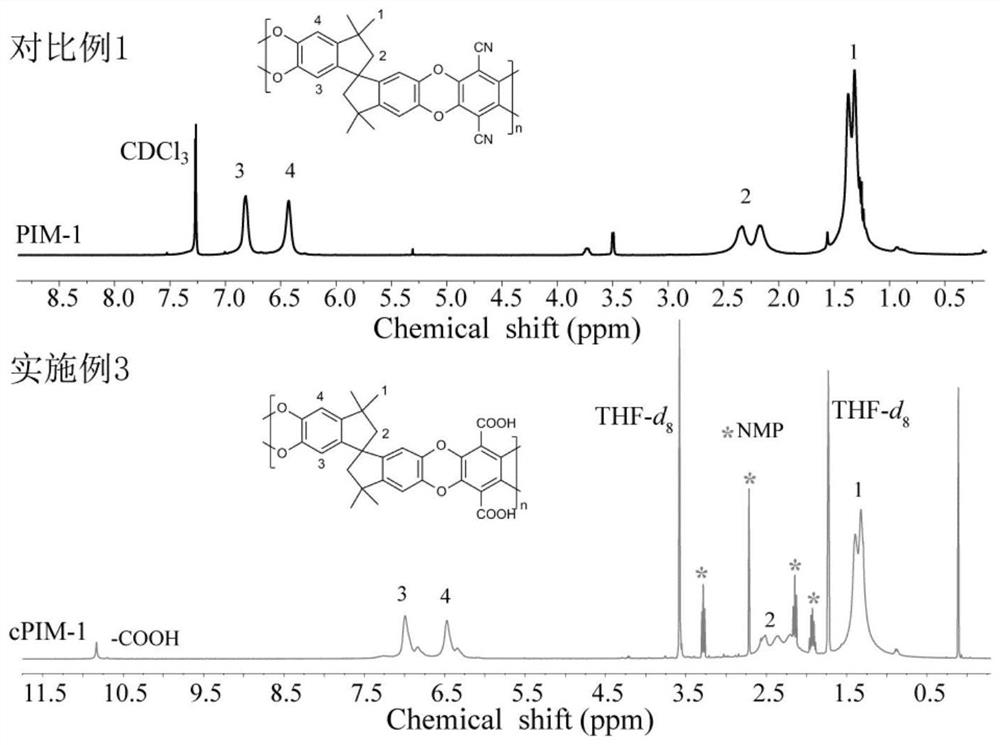
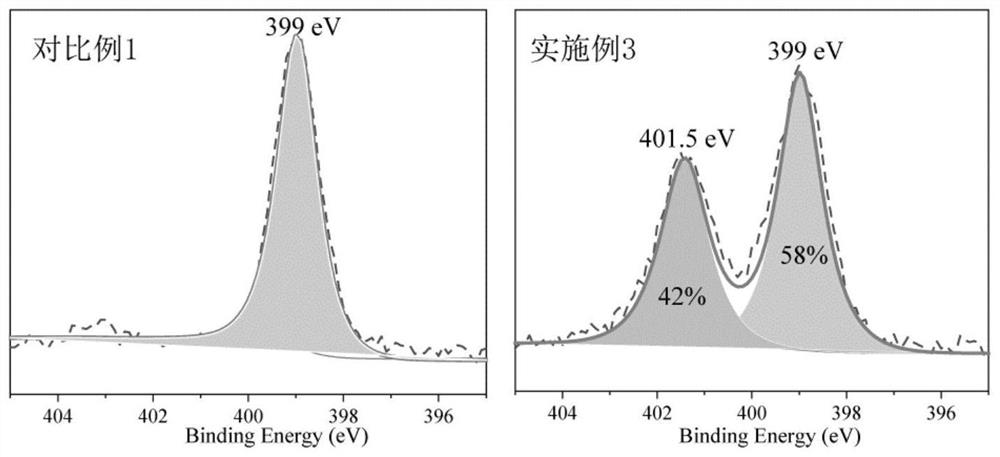
[0039] Soak the PIM-1 membrane prepared in the second step of Comparative Example 1 in methanesulfonic acid for 10 min, then place the membrane soaked in methanesulfonic acid on a glass plate, and then transfer the glass plate to an oven at 110°C for PIM -1 The hydrolysis and crosslinking reaction of the film, the reaction time is 48h. After the reaction was completed, the membrane was taken out from the oven, soaked and rinsed in deionized water, and dried to obtain a novel membrane cPIM-1 containing carboxyl and triazine groups. The obtained cPIM-1 of embodiment 31 H NMR nuclear magnetic spectrum as figure 1 As shown, the XPS spectrum of nitrogen atom is shown as figure 2 shown. In the cPIM-1 membrane structure prepared in Example 3, 42% of the nitrile groups were crosslinked to form a triazine ring, and the degree of crosslinking of the membrane was 42%.
[0040] The second step, the gas separation performance characterization of cPIM-1: test the cPIM-1 membrane to N 2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com