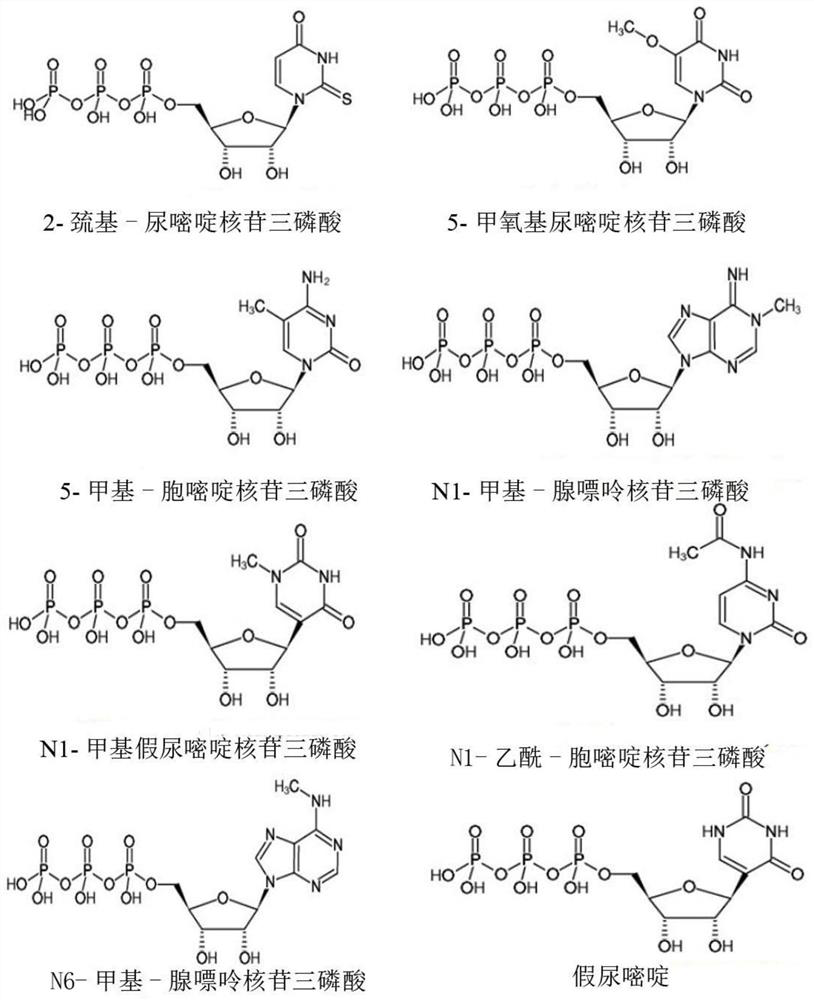
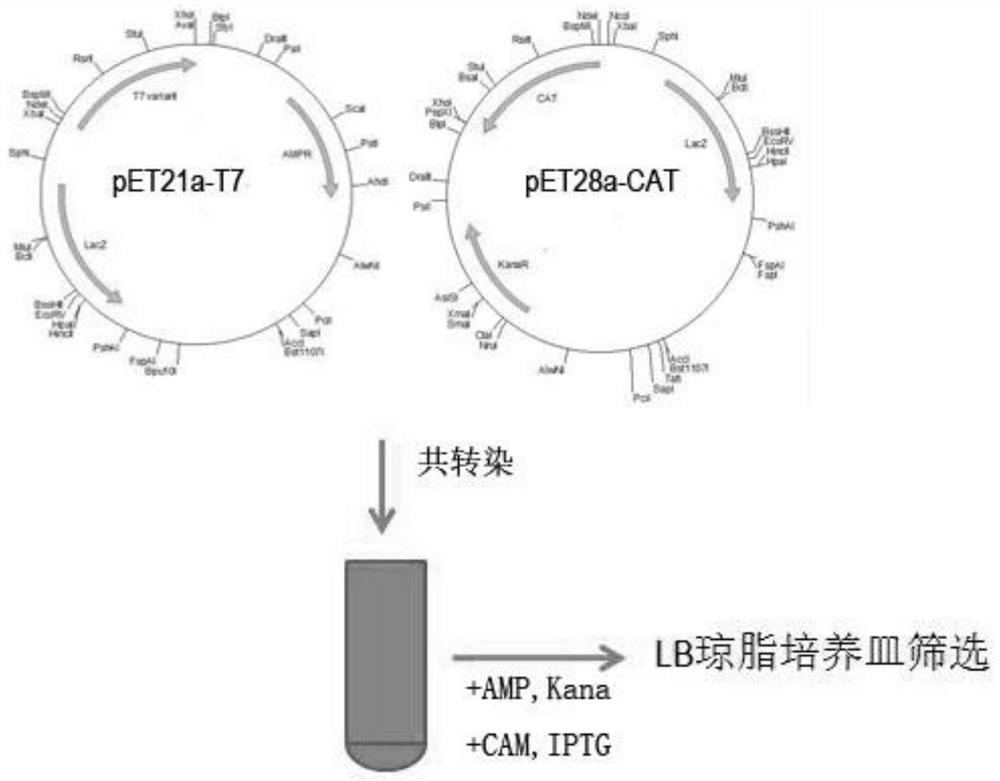
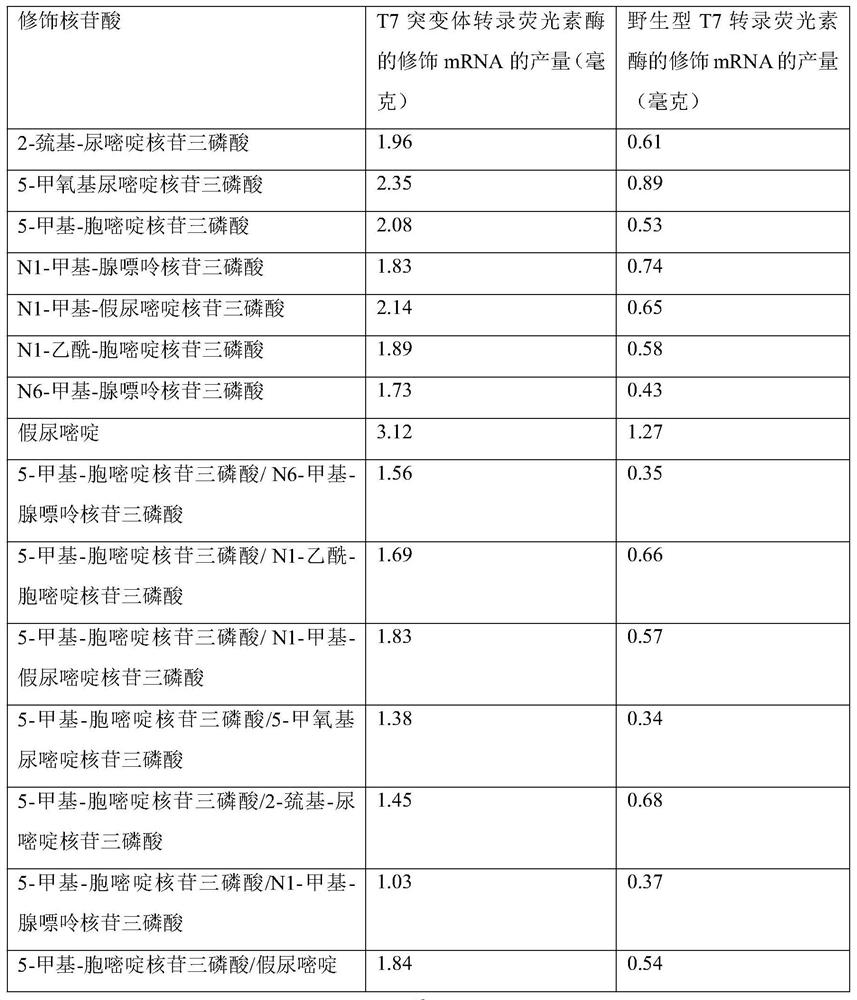
T7RNA polymerase mutant, mRNA, gene, expression vector and cell
A gene expression and polymerase technology, applied in genetic engineering, plant gene improvement, recombinant DNA technology, etc., can solve the problem of transcriptional activity decline
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0056] Example 1: Construction of T7 RNA polymerase mutant screening system
[0057] Dmitry Temiakov et al. used the co-crystallized structure of T7 RNA polymerase and substrate analogs, combined with site-directed mutagenesis technology, to clarify the key mechanism and important amino acid sites of T7 RNA polymerase for substrate selectivity, including Y639, M635, R632, K631, Y571, R627, K472, etc. (Dmitry Temiakov et al., Cell, (2004) 116, 381-391). According to published literature reports such as Ikeda (Ikeda, R.A.et al.Biochemistry, 31:9073-9080,1992 and Ikeda, R.A.etal., Nucl.Acid.Res., 20:2517-2524,1992, JijumonChelliserrykattil etc., Nat Biotechnol.2004 22(9):1155-60.), through the dual-plasmid screening system based on T7 RNA polymerase on the transcriptional activity of T7 promoter, the functional screening of T7 RNA polymerase mutants can be realized. The present invention adopts the same principle and constructs a dual-plasmid screening system for screening T7 RN...
example 2
[0068] Example 2: Protein expression, purification and preliminary functional identification of T7 RNA polymerase mutants
[0069] Since the mutant library of the T7-encoding gene is constructed for amino acids that are key to substrate selectivity, the T7 RNA polymerase mutant obtained through the preliminary screening of Example 1 above is considered to have the ability to utilize modified nucleotides to transcribe Potential for synthetically modified mRNA. By expanding the culture of the T7 RNA polymerase mutant strains screened above, and using the 6xHis tag attached to the C-terminus of the pET21a(+) vector, based on the affinity between the His tag and the Ni-NTA Agarose purification medium, the T7 RNA Polymerase mutant proteins were purified. The specific steps are as follows: collect the bacteria by centrifugation (9000rpm, 10 minutes of centrifugation), resuspend the bacteria in lysis buffer (50mM Tris, pH=8.0, 50mM NaCl), sonicate the bacteria, and centrifuge (12000...
example 3
[0070] Example 3: T7 RNA polymerase mutants use modified nucleotides to transcribe and synthesize luciferase modified mRNA, and compare the transcriptional activity with wild-type T7 RNA polymerase
[0071]The T7 promoter on pET28a was used to test the transcriptional activity of the T7 RNA polymerase mutant by linking the luciferase gene shown in SEQ ID No.6. According to reports by Gallie G.R et al. (Gallie G.R et al., Gene, 1999,165,233-238), the 5' untranslated sequence (5' UTR of tobacco mosaic virus (TEV), its DNA sequence is shown in SEQ ID No.6) Can be used to mediate post-transcriptional translation. Therefore, the 5'UTR of TEV is used here to mediate the translation of luciferase. TEV 5'UTR is shown in SEQ ID No:7 to SEQ ID No:8. In addition, a 100nt poly(dT) was introduced into the transcription template, and a 100nt poly(A) tail structure was obtained through complementary pairing during the transcription process. The 5'UTR, luciferase gene and 100 nt poly(dT) D...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com