A nano-upgrade automatic detection method for genotyping 1536 endpoints
A technology for genotyping and detection methods, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of low sample DNA yield, incompatibility of fluorescence detection equipment, and large amount of DNA used, and achieves the application of The effect of wide platform and lower probe synthesis cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
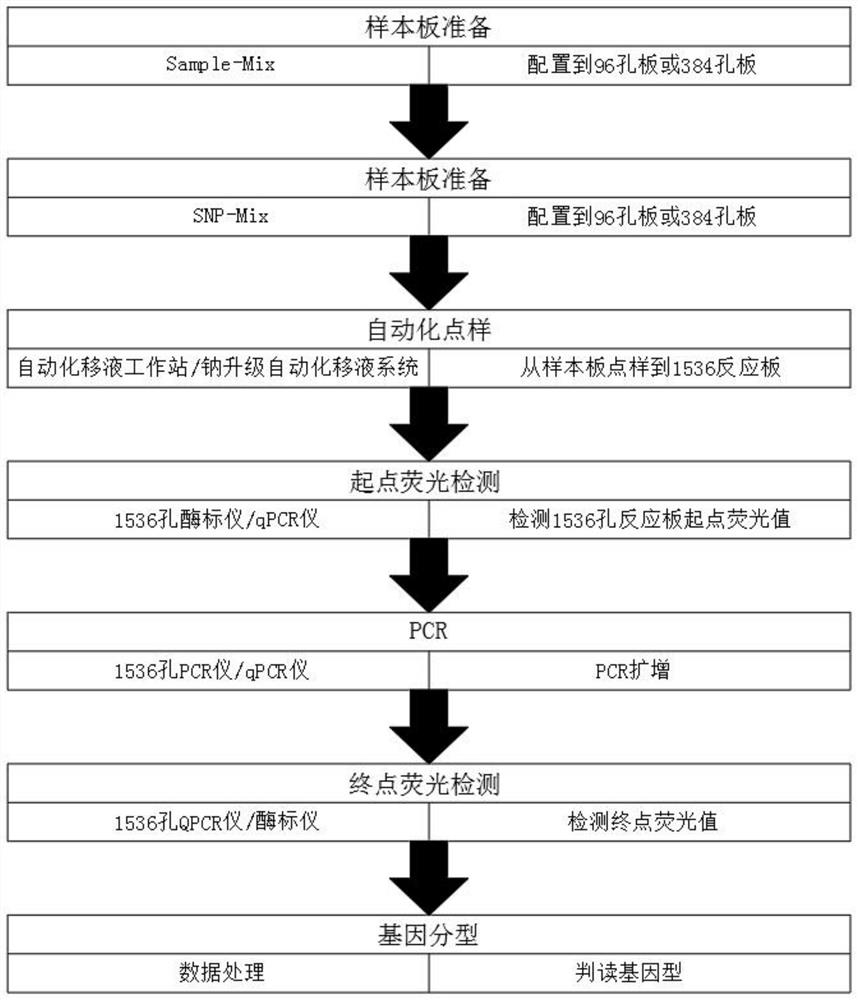
[0045] Such as Figure 1-3 As shown, the end-point ultra-high-throughput automated genotyping detection method based on the enzyme-labeled system:
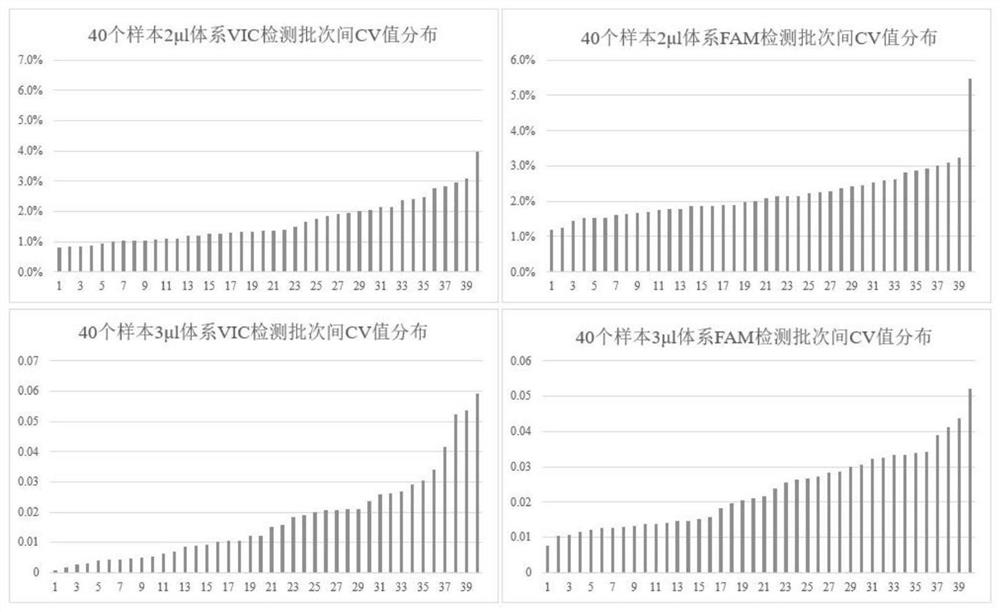
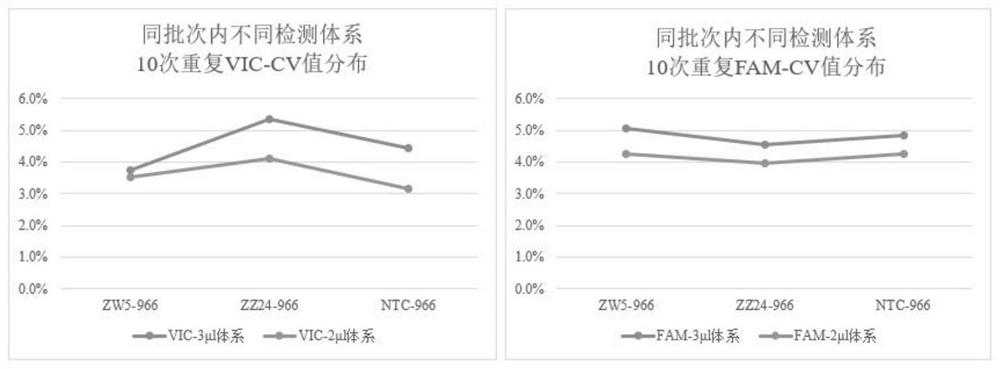
[0046] Using the nucleic acid samples of 10 male fish (ZZ genotype) and 8 female fish (ZW genotype) of known gender, two SNP sites 8935966 (abbreviated as 966) and 8936186 (abbreviated as 186), the method was carried out. Validation of method genotyping accuracy. Two of the loci are: male homozygous genotype, corresponding to VIC signal; female heterozygous genotype, corresponding to VIC and FAM signal:
[0047] S1. Sample plate preparation:
[0048] One SNP site reaction system for one sample in the sample plate: 2×PARMS master mix 0.5 μl, DNA template (50ng / μl) 0.4 μl, ddH2O 0.1 μl; prepare the Sample for each sample according to the number of SNP sites required for each sample -Mix, dispense to 96-well plate;
[0049] S2, SNP plate preparation:
[0050] One SNP site reaction system for one sample in the SNP plate: 2×PARMS ...
Embodiment 2
[0086] Such as figure 1 , 4 As shown, the ultra-high-throughput automated genotyping detection method based on the qPCR system:
[0087] Using the nucleic acid samples of 10 male fish (ZZ genotype) and 8 female fish (ZW genotype) of known gender, two SNP sites 8935966 (abbreviated as 966) and 8936186 (abbreviated as 186), the method was carried out. Validation of method genotyping accuracy. The two loci are: male fish homozygous genotype, corresponding to VIC signal; female fish heterozygous genotype, corresponding to VIC and FAM signal.
[0088] S1. Sample plate preparation:
[0089] One SNP site reaction system for one sample in the sample plate: 2×PARMS master mix 0.5 μl, DNA template (50ng / 5 μl) 0.4 μl, ddH2O 0.1 μl; prepare the Sample for each sample according to the number of SNP sites required for each sample -Mix, dispense into 96-well plate or 384-well plate;
[0090] S2, SNP plate preparation:
[0091] One SNP site reaction system for one sample in the SNP plat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com