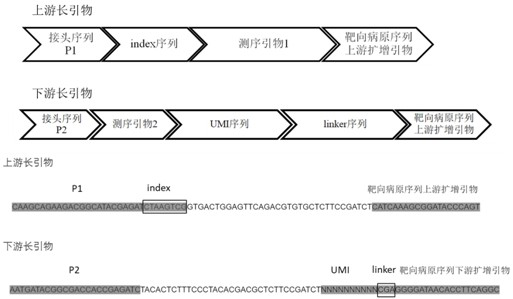
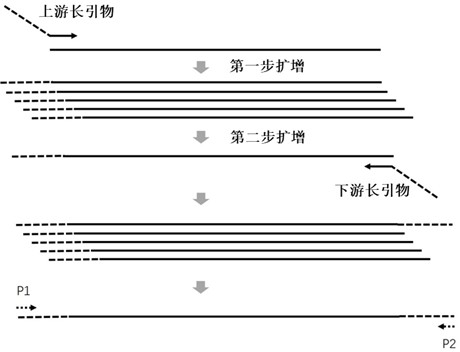
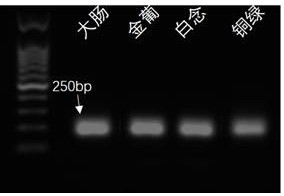
Multi-amplification and high-throughput sequencing method and kit for identifying pathogenic microorganisms
A pathogenic microorganism and multiple amplification technology, which is applied in the field of multiple amplification of pathogenic microorganism identification combined with high-throughput sequencing methods and kits, can solve the problems of cumbersome operation, increase of patient medical expenses, cumbersomeness, etc., and improve the efficiency of library construction , shorten the detection cycle, and simplify the operation process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] Embodiment 1 The flow process of pathogenic microorganism identification of the present invention
[0071] Method of the present invention specifically comprises the steps:
[0072] 1. Sample processing:
[0073] (1) Collection and transportation: collect clinical samples from patients, and adopt different collection and transportation methods according to different sample types.
[0074] (2) Pretreatment: According to the requirements of the sample type, pre-extraction treatment is carried out. If it is a sample of alveolar lavage fluid, it needs to be treated with wall breaking, and if it is a blood sample, it needs to be treated with plasma separation.
[0075] (3) Nucleic acid extraction: Use the DNA and RNA co-extraction kit developed by Sage (Yuesui Machinery Equipment 20200771), and extract DNA / RNA according to the instructions. The concentration and purity of the extracted nucleic acid were tested using nanodrop2000 / qubit3.0.
[0076] 2. Before amplification:...
Embodiment 2
[0100] Example 2 Multiple Amplification Reference Strain Research
[0101] 1. Pathogen composition: According to the previously accumulated data and some published literature or research reports, the present invention selects 20 common pathogenic microorganisms of respiratory tract and blood infection as research objects, and the types are shown in the following table:
[0102]
[0103] 2. Nucleic acid preparation of reference strains: 20 standard reference strains of pathogenic microorganisms were purchased from the Guangdong Provincial Culture Collection Center, and the corresponding strains were inoculated on corresponding petri dishes in an ultra-clean bench, and cultured at their respective suitable temperatures for 12-24 h . Pick a single colony, inoculate in 5 ml liquid medium, and culture overnight. Take 1ml of bacterial liquid and use QIAamp DNA MicrobiomeKit to extract nucleic acid according to the instructions. The extracted nucleic acid is tested for concentrat...
Embodiment 3
[0129] Embodiment 3 multiple amplification clinical sample research
[0130] In order to illustrate that the sensitivity of the method of the present invention is higher than that of traditional clinical methods, the following comparative experiments were carried out.
[0131] 1. Clinical sample collection:
[0132] The alveolar lavage fluid (BALF) and blood samples of 10 critically ill patients were collected from the ICU of the cooperative hospital. The patients were between 35 and 80 years old, 10 males and 10 males, and the SOFA score was between 1 and 13 points.
[0133] 2. Sample processing:
[0134] All 20 samples were identified using traditional clinical methodologies: BALF and blood culture submission, antibody testing and qPCR testing. Another sample of the same patient carries out the method pre-treatment of the present invention:
[0135] 1) Alveolar lavage fluid: Take 0.5 ml of sample and first pass through a shaker + glass beads to break the wall for 30 minut...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com