Multi-gene combined editing and screening method for microorganisms
A screening method and multi-gene technology, applied in the biological field, can solve the problems of lack of multi-gene combination editing methods, achieve the effect of wide application, fast and efficient multi-gene editing, and reduce workload
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
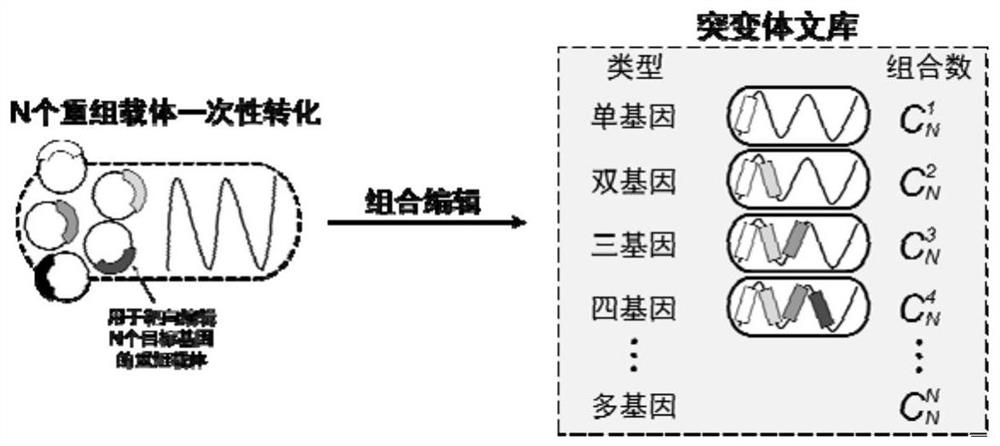
[0055] Combinatorial editing of two genes for one-time transformation with equivalent equivalents matching multiple recombinant vectors
[0056] Combination editing of two histidine kinase genes cac0323 and cac2730 in Clostridium acetobutylicum, theoretically there are three kinds of combination editing of the two genes. According to the sequence information, with the help of software, design appropriate insertion gene sites, respectively select insertion sites at 420 / 421s and 172 / 173a, and synthesize IBS, EBS1d, EBS2, and EBS primers corresponding to the respective genes to construct binary genes targeting different genes. Type intron fragments to obtain methylated recombinant vectors pSY6-cac0323 and pSY6-cac2730. Using the electric pulse conversion method, using the initial concentration (OD 600 =0.8) cells to prepare Clostridium acetobutylicum competent, the two recombinant vectors constructed above were equivalently compatible, wherein the recombinant vectors pSY6-cac032...
Embodiment 2
[0058] Combined editing of two genes by one-time transformation of multiple recombinant vectors with editing sites and equivalence matching
[0059] Combination editing of two histidine kinase genes cac0323 and cac2730 in Clostridium acetobutylicum, theoretically there are three kinds of combination editing of the two genes. According to the sequence information, use the software to design the appropriate insertion gene site, select the forward insertion at the 420 / 421s and 192 / 193s sites respectively, and synthesize the IBS, EBS1d, EBS2 and EBS primers corresponding to the respective genes to construct different gene targeting The two-type intron fragments obtained methylated recombinant vectors pSY6-cac0323 and pSY6-cac2730. Using the electric pulse conversion method, using the initial concentration (OD 600=0.8) cells to prepare Clostridium acetobutylicum competent, the two recombinant vectors constructed above are equivalently compatible, the volume of the recombinant vect...
Embodiment 3
[0061] Combinatorial editing of three genes in one-time transformation with equivalent equivalents matching multiple recombinant vectors
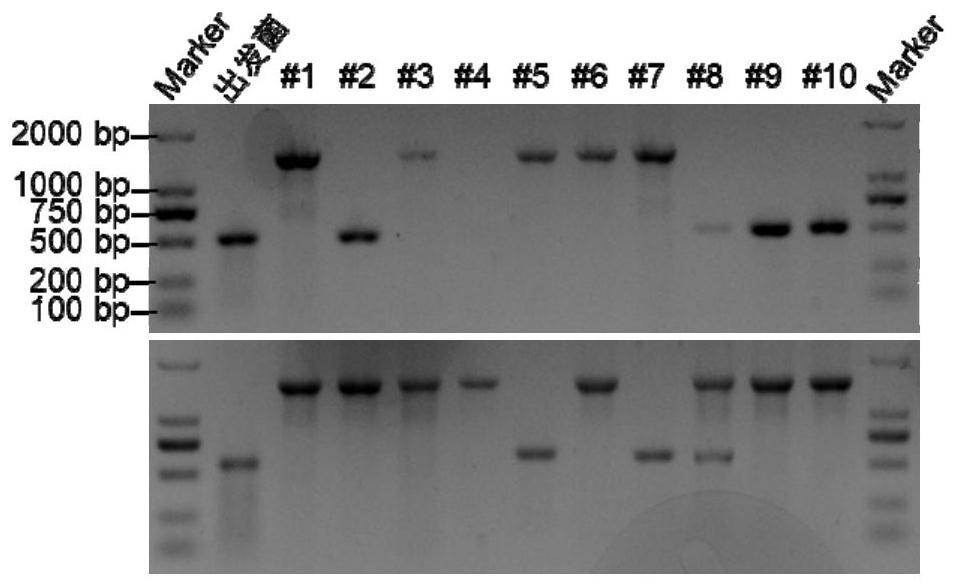
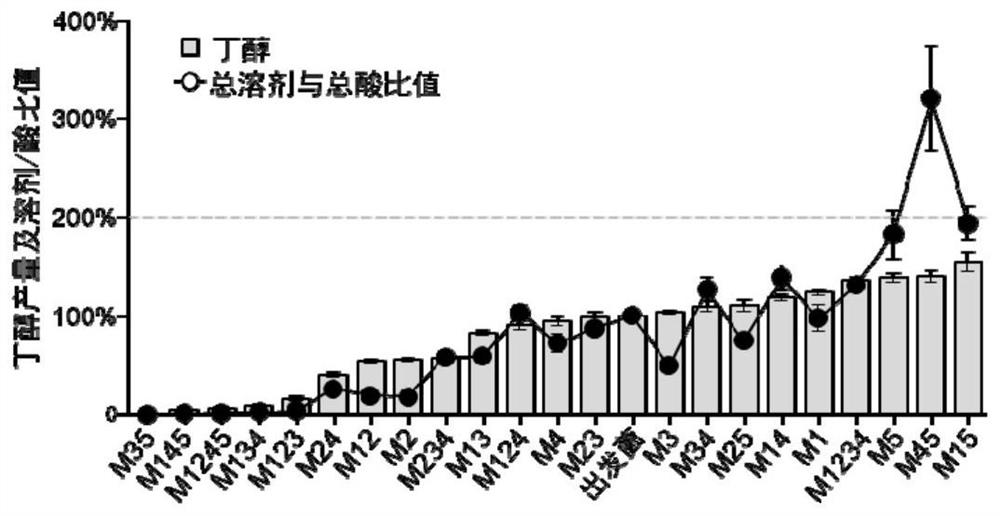
[0062] Combinatorial editing of three genes in the central carbon metabolism pathway in Clostridium acetobutylicum: lactate dehydrogenase gene ldh (cac0267), phosphate transacetylase gene pta (cac1742) and butyrate kinase gene bukII (cac1660), theoretically three There are 7 types of combinatorial editing for each gene. According to the sequence information, with the help of software, design appropriate insertion gene sites, select the forward insertion sites as 171 / 172, 81 / 82, and 421 / 422, and synthesize IBS, EBS1d, EBS2, and EBS primers corresponding to the respective genes to construct Targeting the type II intron fragments of different genes, the methylated recombinant vectors pSY6-cac0267, pSY6-cac1742 and pSY6-cac1660 were obtained. Using the electric pulse conversion method, using the initial concentration (OD 600 =0.8) cells to pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com