A genome editing method based on the endogenous CRISPR-Cas system of Zymomonas mobilis and its application
A Zymomonas and genome editing technology, applied in the field of genetic engineering, can solve problems such as toxicity, nuclease inactivity, host large cells, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 Construction of Zymomonas mobilis Endogenous CRISPR-Cas Genome Editing System
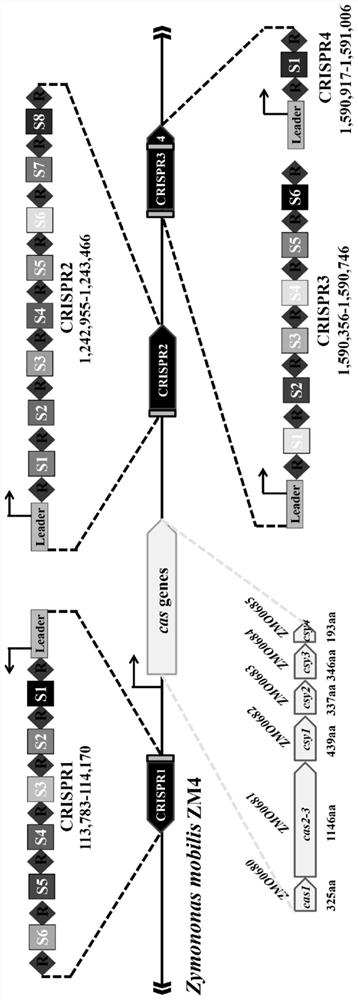
[0040] (1) CRISPR-Cas phylogenetic analysis of the genome encoding of Zymomonas mobilis
[0041] The premise of the present invention is that the research strain needs to encode the CRISPR-Cas system itself and have DNA splicing activity, which requires the host genome to encode the CRISPR cluster and the complete Cas protein system.
[0042]Taking Zymomonas mobilis Z.mobilis 4 as the model strain, the sequencing data of Z.mobilis 4 were analyzed. The results showed that the genome of the strain encoded four CRISPR structural sequences. According to their sequence in the genome, we sequenced them named CRISPR1-CRISPR4, see figure 1 . CRISPR1 occupies the 113,783-114,170 region of the genome and contains 7 repeat sequences; CRISPR2 occupies the 1,244,355-1,245,866 region and contains 9 repeat sequences; CRISPR3 occupies the 1,598,754-1,599,144 region and contains 7 repeat sequence...
Embodiment 2
[0074] Example 2 Application of Genome Editing Method Based on Zymomonas mobilis Endogenous CRISPR-Cas System in Gene Knockout
[0075] In this embodiment, the selected target gene is the ZMO0038 gene encoded by the Zymomonas mobilis genome, and the specific steps are as follows:
[0076] (1) GuideRNA sequence selection: The 32 bp sequence immediately downstream of any 5'-CCC-3' was intercepted from the target gene sequence ZMO0038 as a guideRNA, and the sequence could be located on any strand of the genome.
[0077] ZMO0038-guideRNA primer sequence:
[0078] 0038-gRNA1-F: GAAAATGCCGCTTTCGATCAGGCCGCCGCCAAGATT, see SEQ ID NO:9;
[0079] 0038-gRNA1-R: GAACAATCTTGGCGGCGGCCTGATCGAAAGCGGCAT, see SEQ ID NO: 10;
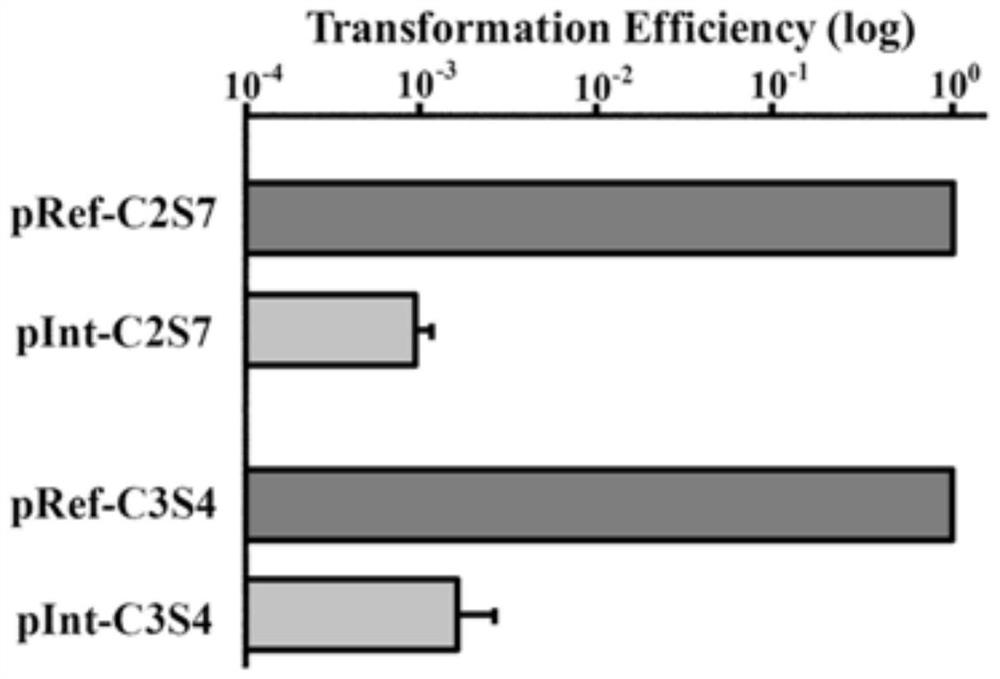
[0080] (2) guideRNA primer sequence is constructed on the vector, see Figure 5 : The above-mentioned plasmid containing the artificial CRISPR expression unit was linearized with BsaI (Table 5), annealed with the guideRNA primer (same as above, Table 2) and ligated with ...
Embodiment 3
[0103] Example 3 Application of Genome Editing Method Based on Zymomonas mobilis Endogenous CRISPR-Cas System in Site-directed Insertion of DNA Sequence
[0104] In this embodiment, a His-Tag tag is inserted at a fixed point after the start code ATG of the target gene ZMO0038, see Figure 8 , so that the encoded protein can be purified through a nickel column. This method can be extended to any protein to be purified. Specific steps are as follows:
[0105] (1) ZMO0038(His-Tag)-guideRNA sequence selection: intercept the 32 bp sequence immediately downstream of the 5'-TCC-3' near the start codon ATG of the target gene sequence ZMO0038 as a guideRNA, and this sequence can be located on any strand of the genome.
[0106] ZMO0038(His-Tag)-guideRNA primer sequence:
[0107] 0038His-gRNA-F: GAAAATCTTGACTCCCTCCATGCACTTAAAAAATCT, see SEQ ID NO: 18;
[0108] 0038 His-gRNA-R: GAACAGATTTTTTAAGTGCATGGAGGGAGTCAAGAT, see SEQ ID NO:19.
[0109] (2) GuideRNA is constructed on the vector:...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com