FMR1 (fragile x mental retardation 1) gene CGG repetition number and methylation detection kit and detection method
A detection kit and repeat number technology are applied in the field of FMR1 gene CGG repeat number and methylation detection kits, which can solve the problems of inaccurate detection results and achieve the effects of accurate interpretation, easy clinical application and promotion, and simple operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
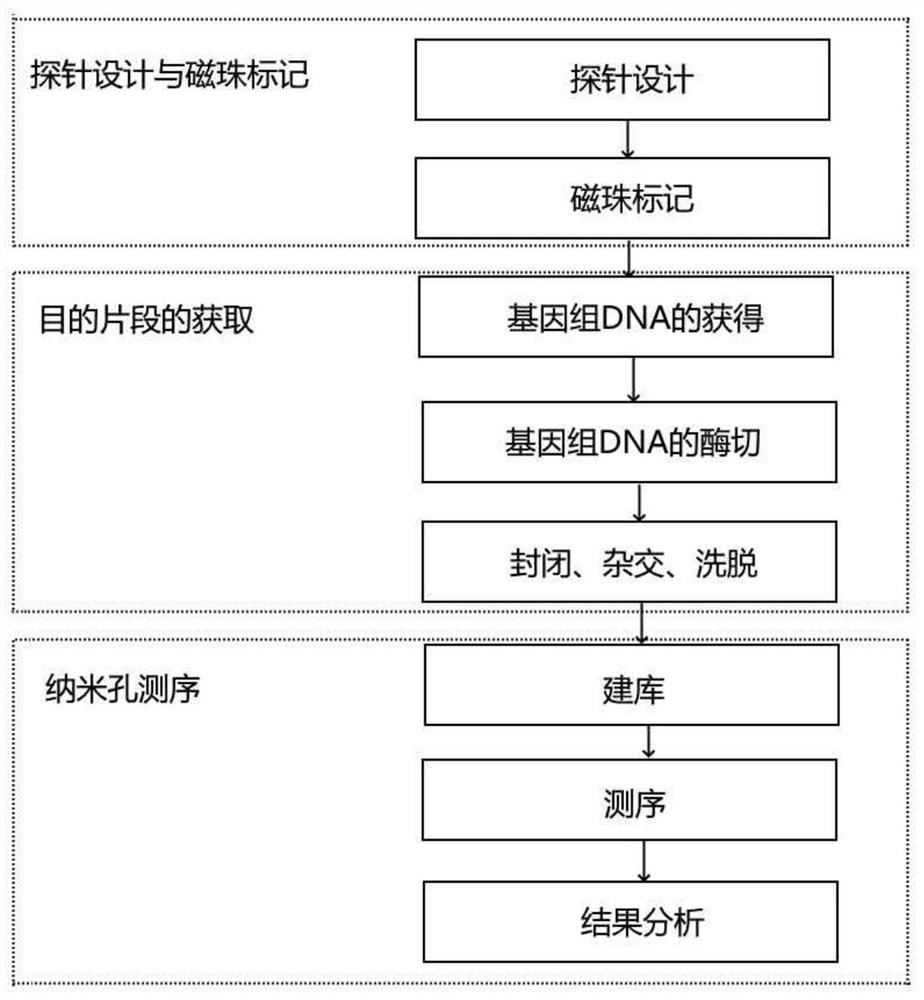
[0060] Example 1. Magnetic bead-labeled capture probes
[0061] The process for magnetic bead labeling of capture probes is as follows:
[0062] (1) Take two test tubes, add 20-100 μl (preferably 30 μl) of amino-modified magnetic beads (20g / L) solution respectively, and discard the supernatant;
[0063] (2) Add 100 μl of cross-linking agent (5%) to two test tubes, shake and mix once every 5-20 minutes (preferably 10 minutes) at room temperature, and shake and mix for 2-5 times (preferably 3 times) ), discard the supernatant;
[0064] (3) Add phosphate buffer solution (pH = 7-8, preferably pH = 7.4) to the two test tubes, shake and wash the magnetic beads, and discard the supernatant;
[0065] (4) Add F10 and R10 to the two test tubes, shake and mix once every 5-20 minutes (preferably 10 minutes), for a total of 2-8 times (preferably 5 times).
[0066] For the method of modifying the magnetic beads with amino groups, please refer to the prior art. The crosslinking agent may...
Embodiment 2
[0067] Embodiment 2. Acquisition of Target Fragment
[0068] The samples used in the detection kit and detection method of the present invention may be whole blood samples, dried blood spot samples, or oral epithelial cell samples. The above-mentioned samples can be extracted through corresponding extraction kits according to the instructions of the kits.
[0069] For the reaction system and reaction conditions of genomic DNA digestion, please refer to the prior art. Briefly, the digestion reaction process of genomic DNA is as follows:
[0070] (1) Take an appropriate amount of sample genomic DNA;
[0071] (2) Add PmlI, NaeI and Buffer to the reaction system according to the instructions;
[0072] (3) Carry out the enzyme digestion reaction at 37°C, and refer to the instruction manual for the reaction time;
[0073] (4) Store the digested product at 4°C for subsequent experiments.
Embodiment 3
[0074] Example 3. Capture Probes Capture Target Fragments
[0075] The process of capturing the target fragment by the capture probe includes pre-hybridization, blocking, hybridization and elution.
[0076] One), the process of pre-hybridization and sealing is as follows:
[0077] (1) Add 50-100 μl (preferably 60 μl) of prehybridization solution to the F10 capture probe and the R10 capture probe coupled to magnetic beads, and then add an equal volume of the blocking solution, and shake for 0.5 ~ 2 hours (preferably 1 hour);
[0078] (2) adding the phosphate buffer solution to wash;
[0079] (3) Add 100 μl of hybridization solution (Tris-HCl buffer, PH=7.4), and add the digested DNA sample, heat at 95°C for 5 minutes to decompose the DNA into single-stranded molecules, and shake the hybridization reaction at 40°C for 15~ 60 minutes (preferably 30 minutes);
[0080] (4) Discard the supernatant, add double distilled water to shake and rinse for 2 minutes, repeat three times; ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com