Compound function prediction method based on neural network and connection graph algorithm
A neural network and prediction method technology, applied in the field of drug function evaluation based on neural network and gene enrichment algorithm, can solve problems such as prediction, and achieve the effect of accelerating the drug development process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0027] The present invention is further described below.
[0028] Antioxidant compound function prediction based on neural network and connection graph algorithm, including the following steps:
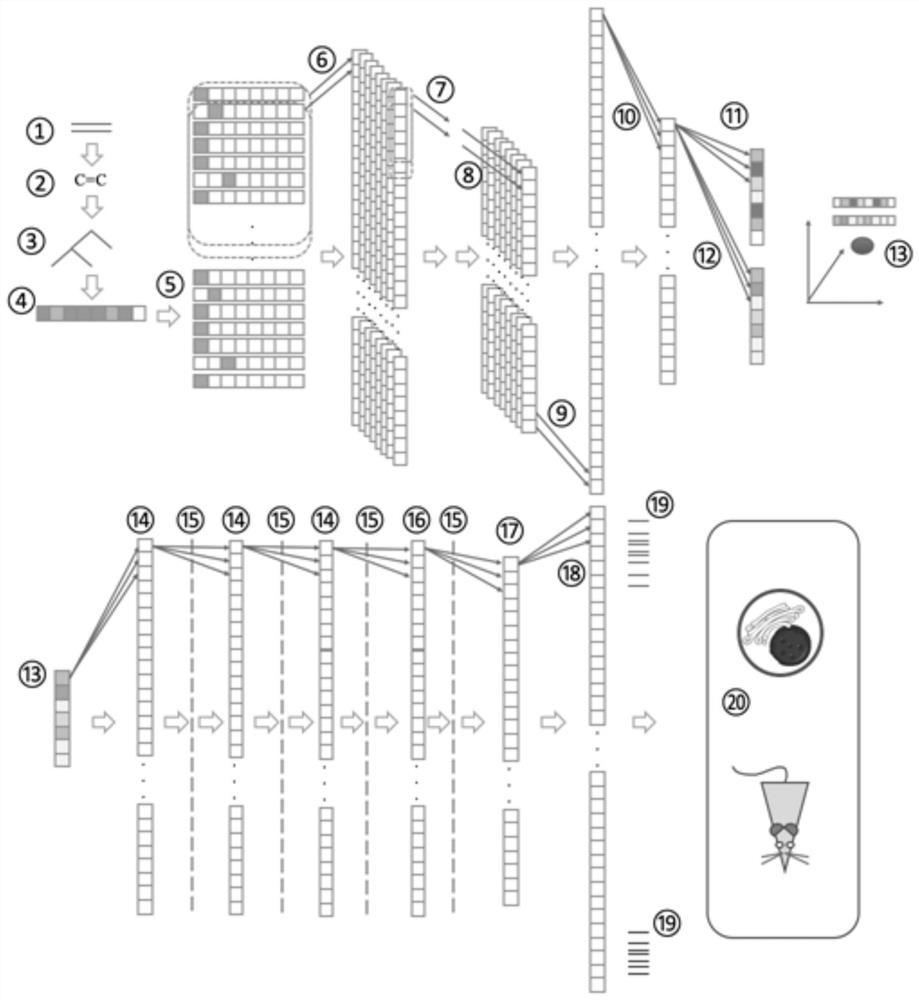
[0029] (1) Obtain the molecular formula of the compound from the public database, construct the neural network of the compound molecular change autoencoder (VAE), train the neural network based on the change of the molecular formula of the compound, and the output of the neural network is the encoding vector of the compound molecule;
[0030] For example, obtain compound molecular formula from public databases PubChem, ensemble and zinc, construct compound molecule-encoding vector neural network according to molecular structure, and train self-encoding neural network (GrammarVAE) based on the change of compound molecular formula syntax, see figure 1 , the output is the encoding vector of the compound molecule;
[0031] (2) Obtain the compound-gene expression change data from the publ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com