Method for constructing 14-3-3 epsilon gene knockout cell strain based on CRSIPR technology and application of 14-3-3 epsilon gene knockout cell strain
A gene knockout, cell line technology, applied in applications, animal cells, genetic engineering, etc., can solve the problems of HP-PRRSV replication increase, and achieve the effect of short cycle, low cost and stable cell line
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
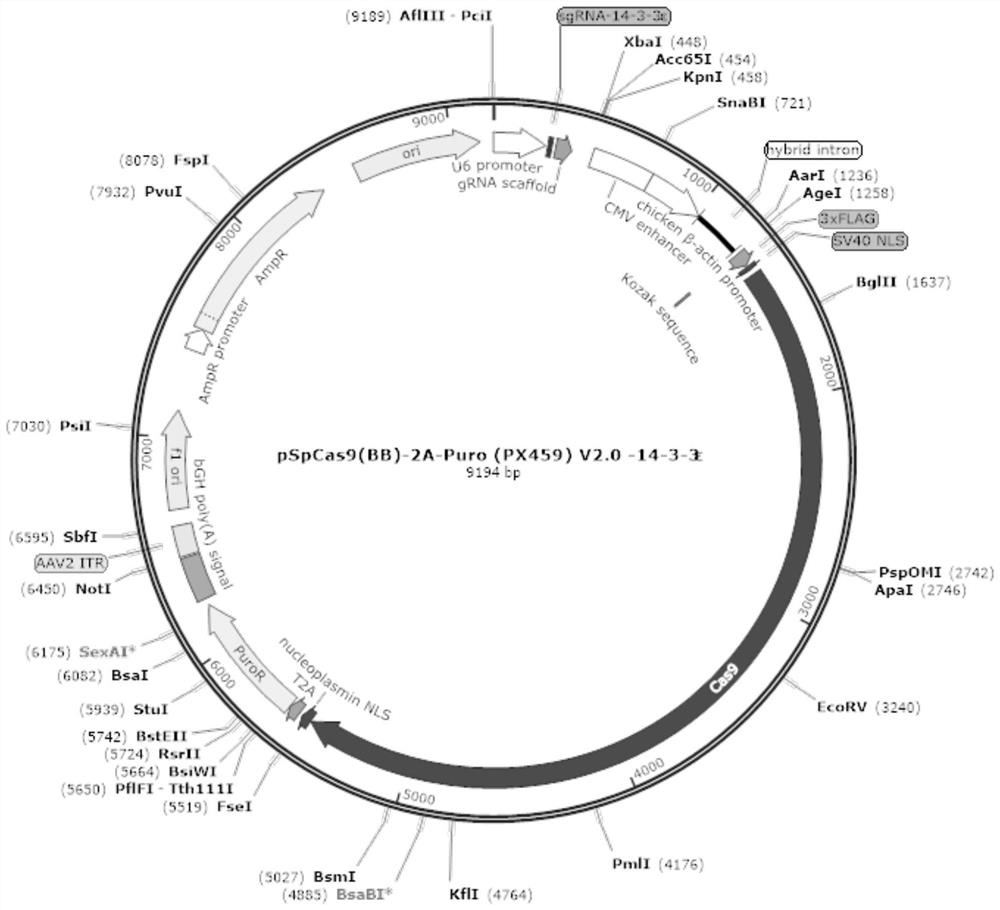
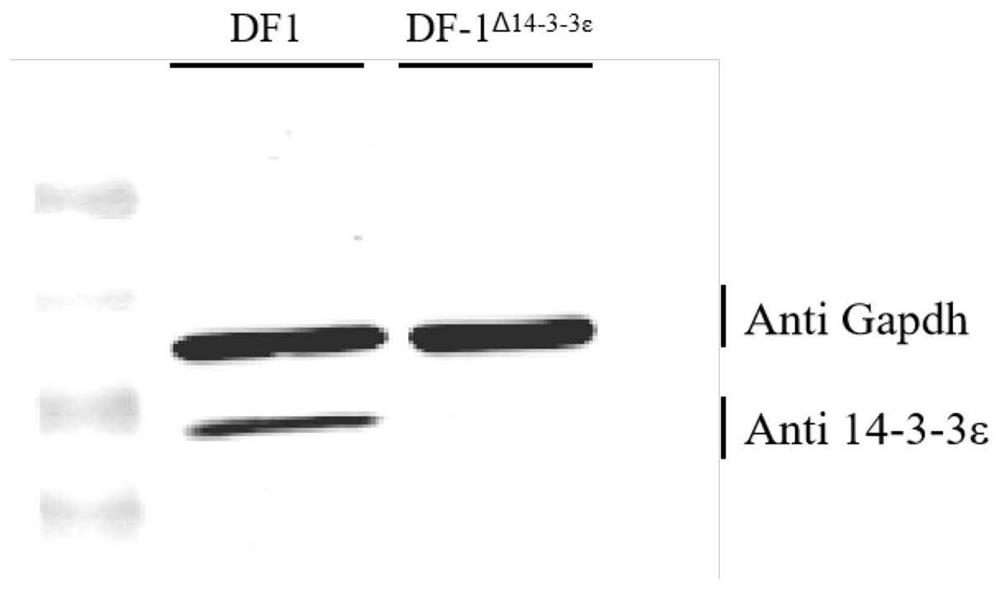
[0028] Example 1 Construction of 14-3-3ε gene knockout cell line based on CRSIPR technology
[0029] 1. sgRNA design and synthesis
[0030] Download the corresponding nucleotide sequence according to the chicken source 14-3-3ε (ENSGALG00000002661) gene published by ensembl, and use the online sgRNA design tool (http: / / crispor.tefor.net) in the second exon region of 14-3-3ε ) designed 14-3-3ε sgRNA (small guide RNA), and the primers were sent to Sangon Bioengineering (Shanghai) Co., Ltd. for synthesis. The synthetic primer sequences are as follows:
[0031] sgRNA-F: 5'-caccgGGTTGAATCAATGAAGAAAG-3';
[0032] sgRNA-R: 5'-aaacCTTTCTTCATTGATTCAACCc-3'.
[0033] 2. Construction of recombinant eukaryotic expression plasmid pX459-14-3-3ε
[0034] Annealing: After centrifuging the sgRNA sequence synthesized by the company, add an appropriate amount of ddH 2 O, make the final concentration 100 μmol / L, add T4 ligase after fully dissolving, and use PCR instrument to anneal the sgRNA ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com