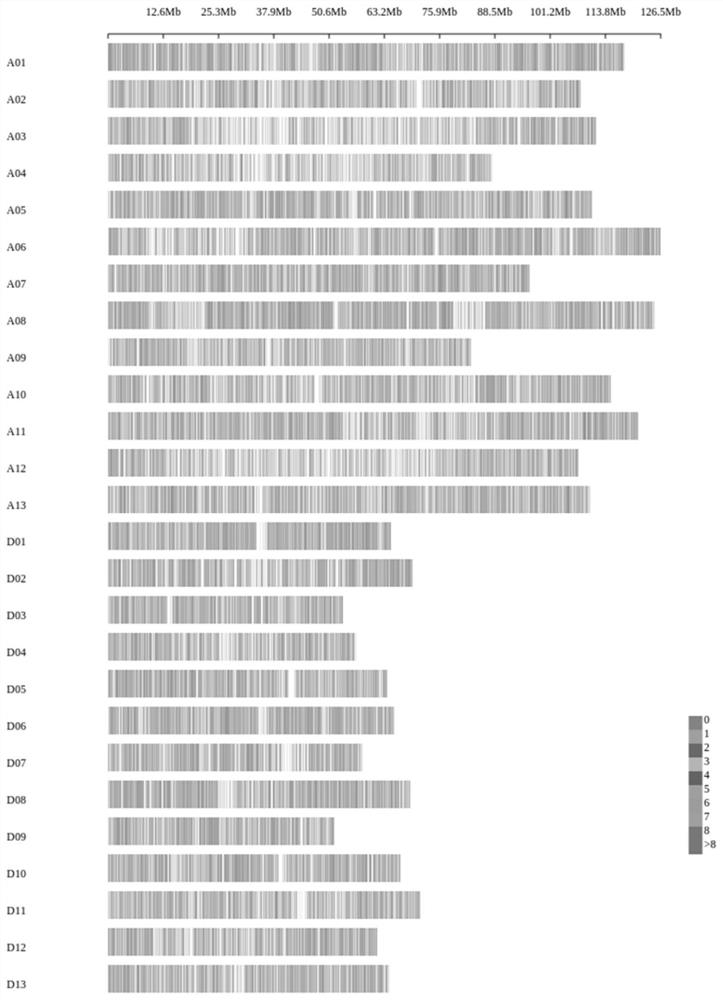
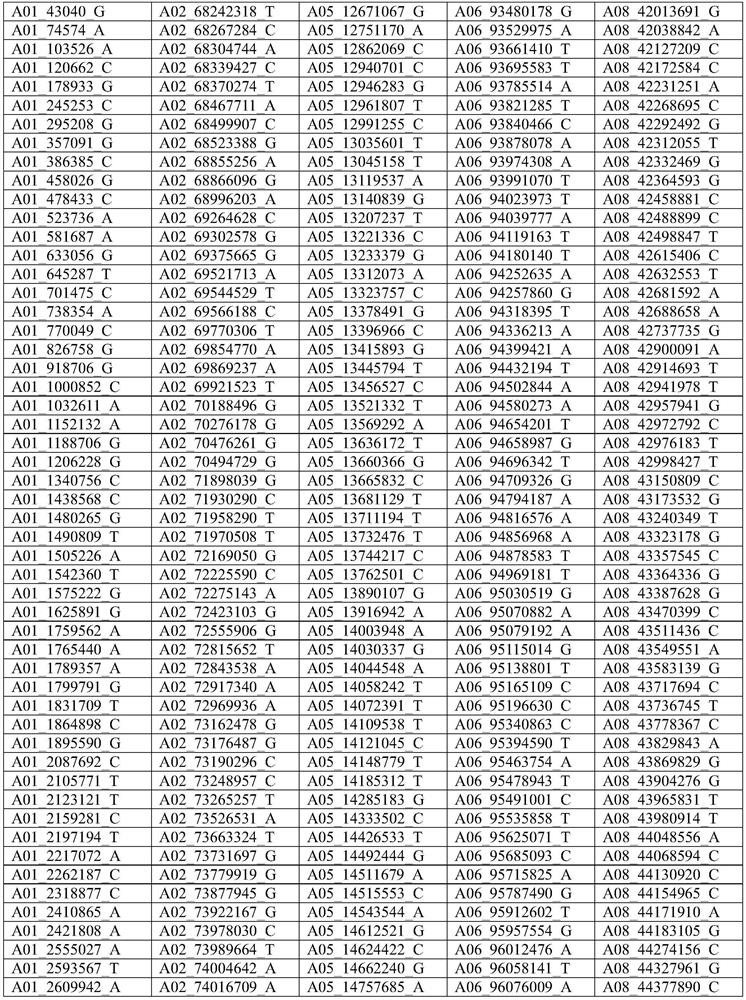
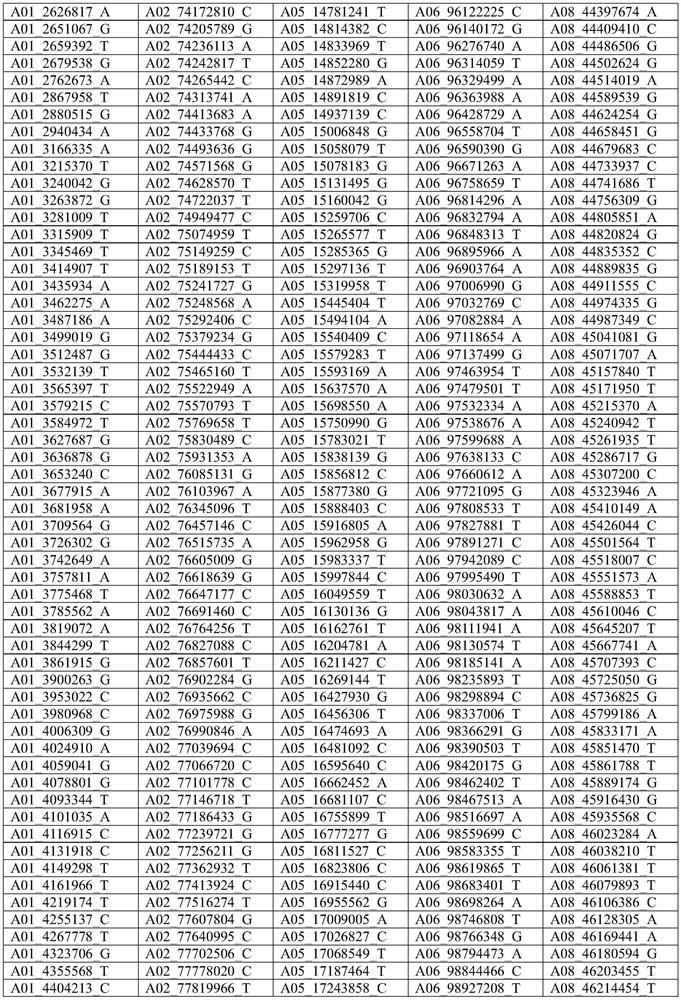
Cotton whole genome 40K mononucleotide site, and application thereof in cotton genotyping
A genome-wide, single-nucleotide technology, applied in the fields of genetics, bioinformatics, cotton molecular breeding, and molecular biology, can solve problems such as no chip reports
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1. Cotton 40K single nucleotide loci are used in the preparation of cotton gene chips.
[0029] The specific method for the preparation of the 40K cotton gene chip is: through library construction, hybrid capture, library quality inspection and sequencing analysis, the 40K target loci are finally determined.
[0030] 1. Library Construction of 13 Cotton Varieties
[0031] Add 100 ng of cotton DNA, 4 μL of GenoBaits End Repair Buffer, 3.1 μL of GenoBaits End repair Enzyme to a 0.2 μL PCR tube, and make up 20 μL with water; place the reaction tube in a PCR machine, at 37°C for 20 minutes, and at 72°C for 20 minutes. Then add 2 μL of GenoBaits Ultra DNA ligase, 8 μL of GenoBaits Ultra DNA LigaseBuffer, 2 μL of GenoBaits Adapter, 8 μL of Nuclease-free water, and place in a PCR machine at 22°C for 60 min. Add 1.2 times of GenoPrep DNA Clean Beads, mix thoroughly, place on a magnetic stand, let stand for 3min, and remove the supernatant. Add 100 μL of freshly prepar...
Embodiment 2
[0180] Example 2. Application of cotton 40K single nucleotide liquid-phase gene chip in cotton genotyping.
[0181] The inventor selected a total of 13 tetraploid cotton species materials including the genetic standard line TM-1 of upland cotton, American cotton, semi-wild cotton, the backbone parents of the three cotton regions of the Yangtze River Basin, the Yellow River Basin, and the Northwest Inland of my country. The 40K cotton gene chip prepared in Example 1 detects the genotypes of the 13 variety materials at the 40K single nucleotide locus described in Example 1. The detection rate of 13 kinds of material loci was 98.62~99.96%. By comparing the loci of the reference genome (TM-1) with the loci detected by the chip, the consistency of the detected loci was 99.40%.
[0182] Attached Table 2, 13 Cotton Variety Resources
[0183]
[0184]
[0185] The entire experimental process is completed based on the GenoBaits technology system. The specific experimental proce...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com