Method for detecting diversity of fish species by using environmental DNA technology
A technology of diversity and species, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problem that species cannot be monitored, and achieve the effect of accurate detection results and various types
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] The purpose of this embodiment is to provide a method for detecting fish species diversity using environmental DNA.
[0028] 1. Design primers
[0029] According to the primer design, the sequence of the vertebrate 12S rRNA universal primer is shown in Table 1, and the amplified target band is 106bp. In order to label the sample sequences of 45 sampling points and negative controls (negative control during sampling, negative control during eDNA extraction process, negative control during PCR process), the same specific label was added to the 5' ends of the forward and reverse primers. The tag is designed using OLIGOTAG software, consisting of six nucleotides, and 2 or 3 N bases are added in front of the tag, and the tag tag is shown in Table 2. The 48 sets of 12s rRNA universal primers after adding the tag were sent to Biosynthesis Co., Ltd. for synthesis, and the primer set was obtained. The aforementioned primer set was sequenced. The primer set is the adjusted 12s ...
Embodiment 2
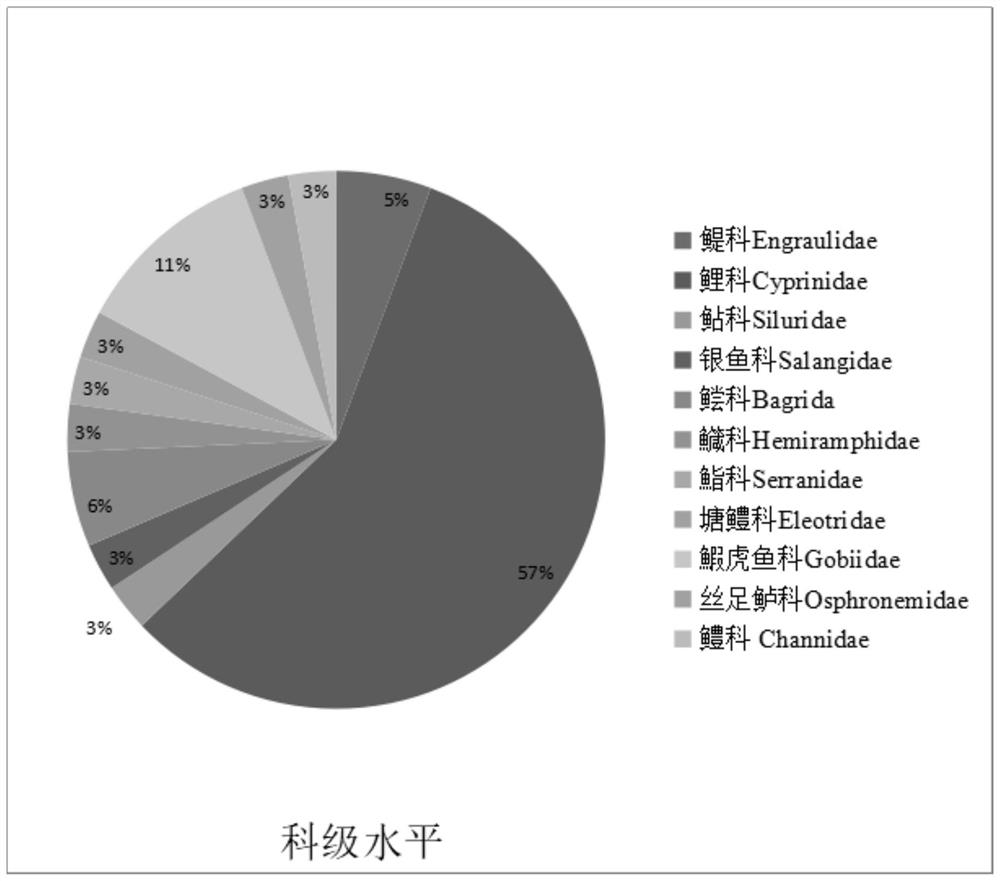
[0045] The purpose of this example is to use the method provided in Example 1 to detect water samples.
[0046] 1. Sampling
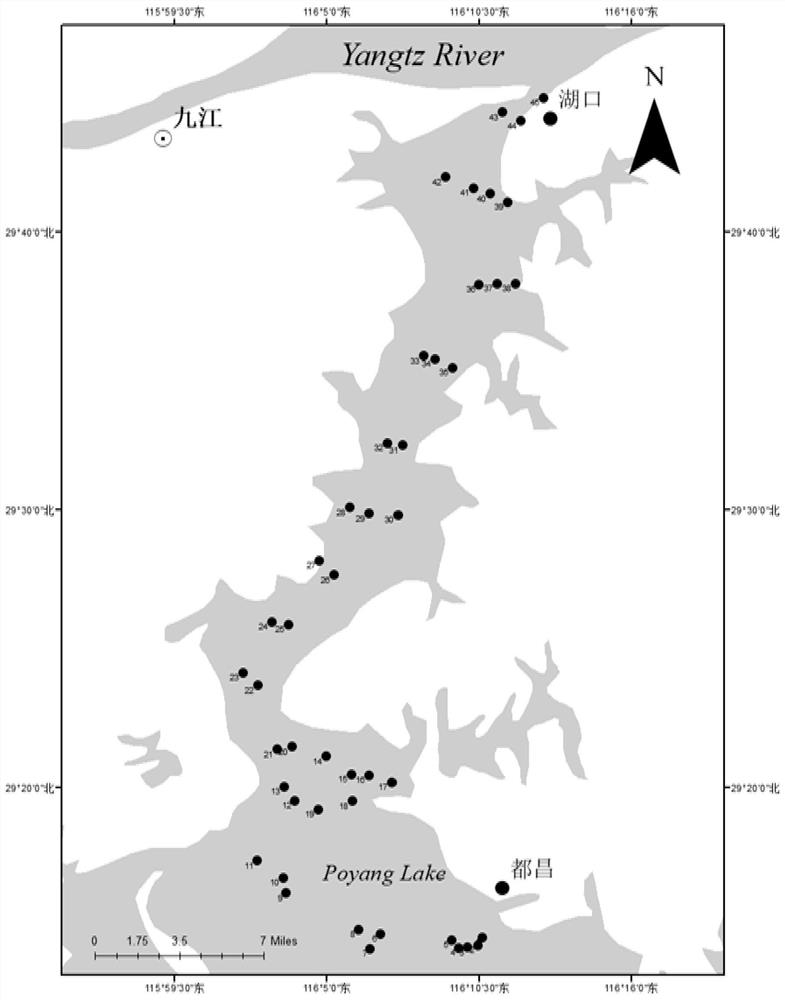
[0047] Adopt according to the method in embodiment 1, altogether 135 samples are arranged at 45 sampling points in Poyang Lake, the sampling point information map is as follows figure 1 shown. Water samples were vacuum filtered using a 0.45 μm mixed cellulose lipid membrane (MCE: Whatman Company) within 24 h. Add a negative control with 1L mineral water as a sample at each testing point to detect whether the sample is contaminated during storage, transportation, and extraction. After each sample was filtered, the filters were placed in 2 mL centrifuge tubes and stored frozen until DNA extraction. Samples from different sampling points are kept in isolation to prevent cross-contamination between samples.
[0048] 2. Filter
[0049] According to the method shown in Example 1, the aforementioned samples were vacuum filtered, and the spare samples were...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com