Primers for detecting SARS-CoV-2 novel coronavirus and kit thereof, detection method and applications
A sars-cov-2 and kit technology, applied in the field of primers for SARS-CoV-2 detection, can solve the problems of 30% to 50%, limiting the wide application of nucleic acid detection, and high false negative rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
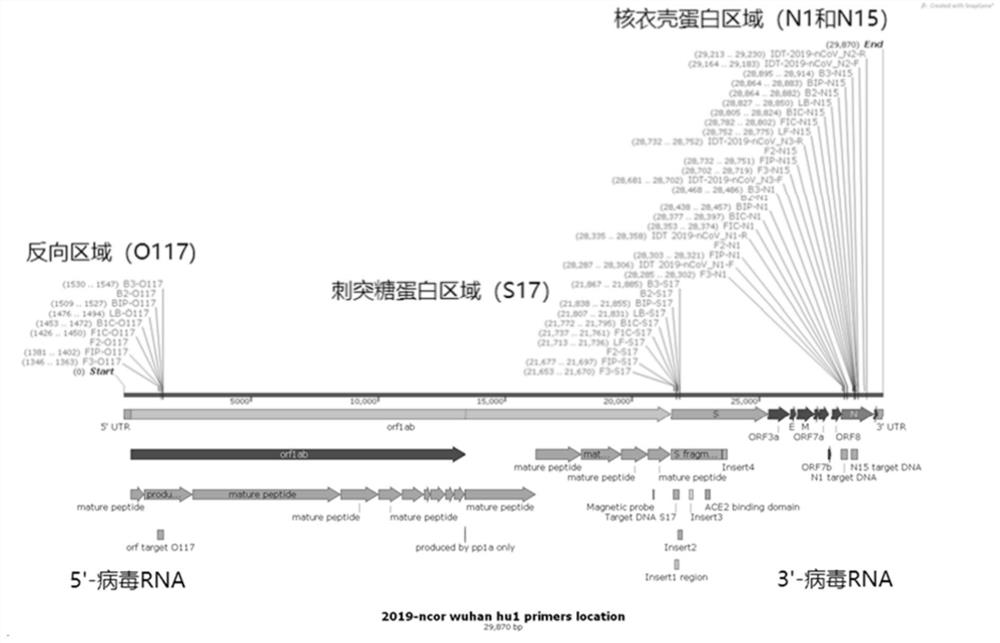
[0045] Example 1. Design of specific primer sequences for detection
[0046] The inventors of the present application designed highly specific primer sequences for the SARS-CoV-2 virus genome sequence (GenBank, NC_045512.2) published in NCBI. In particular, the present application designed 4 primer sets, namely O117, S17, N1 and N15 primer sets, which are specific to different regions in the viral gene sequence. Each primer set contains 6 primers, namely forward primer F3, reverse primer B3, forward inner primer FIP, reverse inner primer BIP, forward loop primer LF and reverse loop primer LB. Primers were designed using the primer design software PrimerExplorer ( http: / / primerexplorer.jp / e / ) as an auxiliary tool.
[0047] The design of the primers in this application takes into account the key characteristics that are highly related to the SARS-CoV-2 virus RNA itself, so as to ensure the high specificity, accuracy and sensitivity of each primer set. SARS-CoV-2 is a single-s...
Embodiment 2
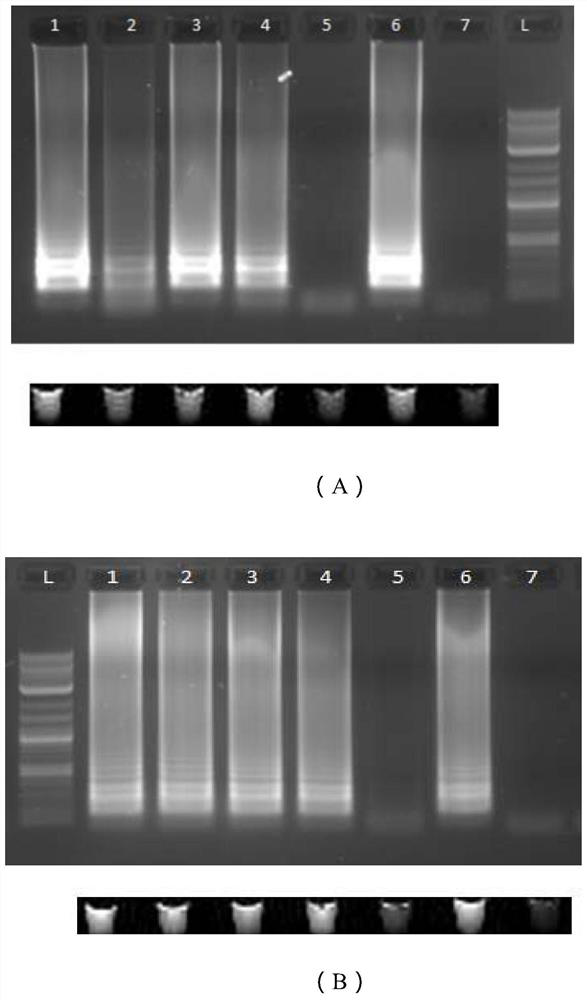
[0055] Example 2: In vitro detection simulation experiment of SARS-CoV-2 virus synthetic DNA fragment target
[0056] This embodiment is used to test the primers of the present application for in vitro (invitro) detection simulation experiments of SARS-CoV-2 synthetic DNA fragments.
[0057] 1. DNA target synthesis and purification
[0058] In order to carry out simulation experiments, DNA fragments targeting four relevant regions of N, S, and Orf1ab target genes were designed, and each fragment contained the T7 promoter for in vitro transcription, namely N1-T7, N15-T7, S17-T7 , O117-T7, its sequence (SEQ ID NO.25-28) is shown in Table 2. Human β-actin primer (Poon LLM, et al.Detection of human influenza A viruses by loop-mediated isothermamplification.Journal of Clinical Microbiology 43,427-430 (2005)) was used as a negative control, and its sequence (SEQ ID NO.29 -32) as shown in Table 3. All designed primers and DNA fragments, as well as the positive control plasmid (201...
Embodiment 3
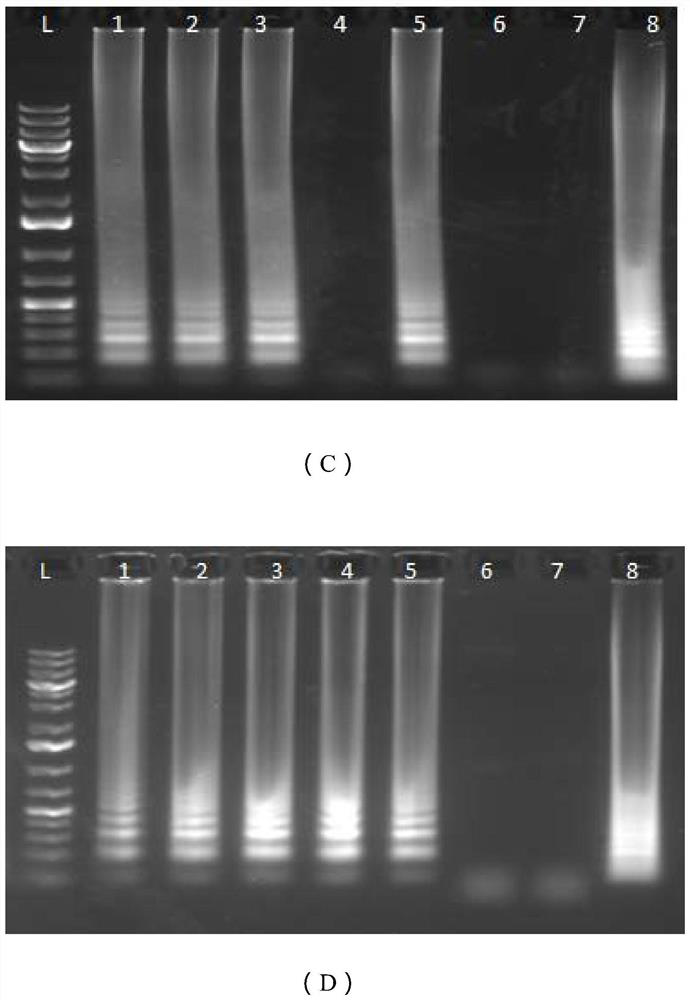
[0083] Example 3: In vitro simulation experiment of SARS-CoV-2 virus RNA target
[0084] This embodiment is an in vitro simulation experiment for testing the primers of the present application for detecting SARS-CoV-2 viral RNA.
[0085] 1. Acquisition and purification of RNA targets
[0086] Use HiScribe TM T7 High Yield RNA Synthesis Kit (HiScribe TM T7 High Yield RNASynthesis Kit, New England Biolabs, UK), the DNA sequence of T7_N, T7_S and T7_O synthesized by IDT described in Example 1 was transcribed in vitro.
[0087] The transcripts were purified using RNeasy Mini Kit (Qiagen, UK), and the concentration and quality of RNA were measured using NanoDrop nucleic acid detector.
[0088] The copy number of each target RNA was determined based on its molecular weight, and diluted to 200,000 copies / μl, 200 copies / μl, 20 copies / μl, and 2 copies / μl for subsequent experiments.
[0089] Using Fast DNA TM The SPIN kit (MP Biomedicals) was used to purify the whole human genome ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com