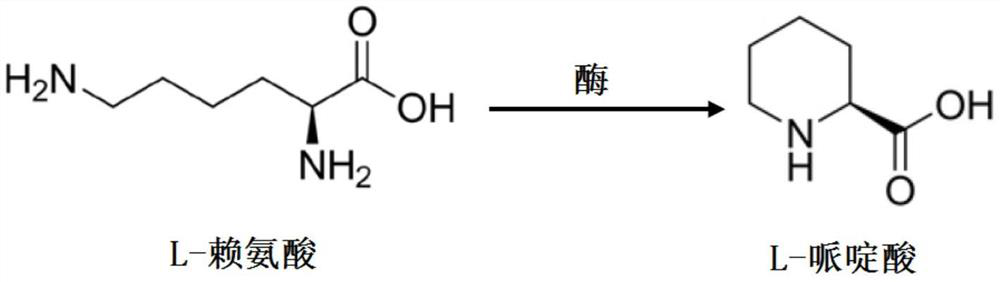
Method for catalytically synthesizing L-piperidine acid by using ornithine cyclization deaminase
A technology of pipecolic acid and ornithine, which is applied in the field of biocatalysis, can solve the problems of low enzyme activity/catalytic efficiency, and great difference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Example 1: Construction of recombinant Escherichia coli expressing wild-type ornithine cyclodeaminase
[0054] For the wild-type ornithine cyclodeaminase SEQ ID NO: 1 derived from Streptomyces melanosporofaciens, an optimized codon sequence SEQ ID NO: 2 suitable for expression in E. The restriction endonuclease sites NdeI and XhoI were designed, and subcloned into the pSH plasmid (including NdeI / XhoI sites) constructed by Zhejiang Huarui Biotechnology Co., Ltd. to obtain the recombinant plasmid pSH-OCD. The recombinant plasmid pSH-OCD was transformed into the expression host Escherichia coli BL21(DE3) to obtain the recombinant Escherichia coli expressing wild-type ornithine cyclizing deaminase (OCD), abbreviated as Ocd.
Embodiment 2
[0055] Embodiment 2: the construction of error-prone PCR and random mutation library
[0056] Using the base sequence of SEQ ID NO:2 as a template, a primer pair Ocd-Nde1-F / Ocd-Xho1-R was designed, and an error-prone PCR technique was used to construct a random mutant library.
[0057] Forward primer Ocd-Nde1-F: 5'-ATATACATATGGAAACCACCGTGC-3',
[0058] Reverse primer Ocd-Xho1-R: 5'-GTCGACTTACTGAAAGCTATACGGG-3'.
[0059] 50μL error-prone PCR reaction system includes: 50ng plasmid template pSH-OCD, 5μL DMSO, 30pmol pair of primers Ocd-Nde1-F and Ocd-Xho1-R, 1X Taq buffer, 0.2mM dGTP, 0.2mM dATP, 1mM dCTP, 1mM dTTP, 7mM MgCl 2 , (0mM, 0.05mM, 0.1mM, 0.15mM, 0.2mM) MnCl 2 , 2.5 units of Taq enzyme (Thermo Fisher Scientific).
[0060] The PCR reaction conditions were: 95°C for 5min; 94°C for 30s, 60°C for 30s, 72°C for 2min / kbp; 30 cycles; 72°C for 10min. The 1kb random mutation fragment was recovered from the gel as a large primer, and MegaPrimer PCR was performed with KOD-pl...
Embodiment 3
[0062] Example 3: Screening of Random Mutant Library
[0063] The transformants in the mutant library were selected and inoculated into a 96-well deep-well culture plate containing 700 μL LB medium containing 100 μg / mL kanamycin, cultured at 37°C for 6 h, and then added with a final concentration of 0.1 mM IPTG, Cool down to 25°C and incubate overnight. Centrifuge at 5000 rpm for 10 min, discard the supernatant, add 200 μL of 50 mM Tris-HCl (pH 7.5), resuspend the bacteria, and use for the determination of OCD enzyme activity.
[0064] The assay protocol for mutant OCD enzyme activity, that is, L-lysine conversion, is as follows:
[0065] Reaction system: 400mM L-lysine, 150mM, pH7.0 potassium phosphate buffer, 0.1% triton X-100, 20mM FeSO 4 , adjust pH to 7.5, bacterial concentration 15v / v%;
[0066] Reaction conditions: 37°C, 150rpm, 24h;
[0067] Termination conditions: 90°C, 5min.
[0068] After the reaction was terminated, it was centrifuged at 13000rpm for 5min, and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com