Quantitative detection method of plant histone variant H3.3 based on MRM
A quantitative detection method and quantitative detection technology, applied in the field of chemical detection, can solve problems such as mutual interference, insufficient ionic strength, modification positioning and quantitative accuracy reduction, etc., to improve the signal-to-noise ratio, reduce chemical background, increase accuracy and The effect of reliability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
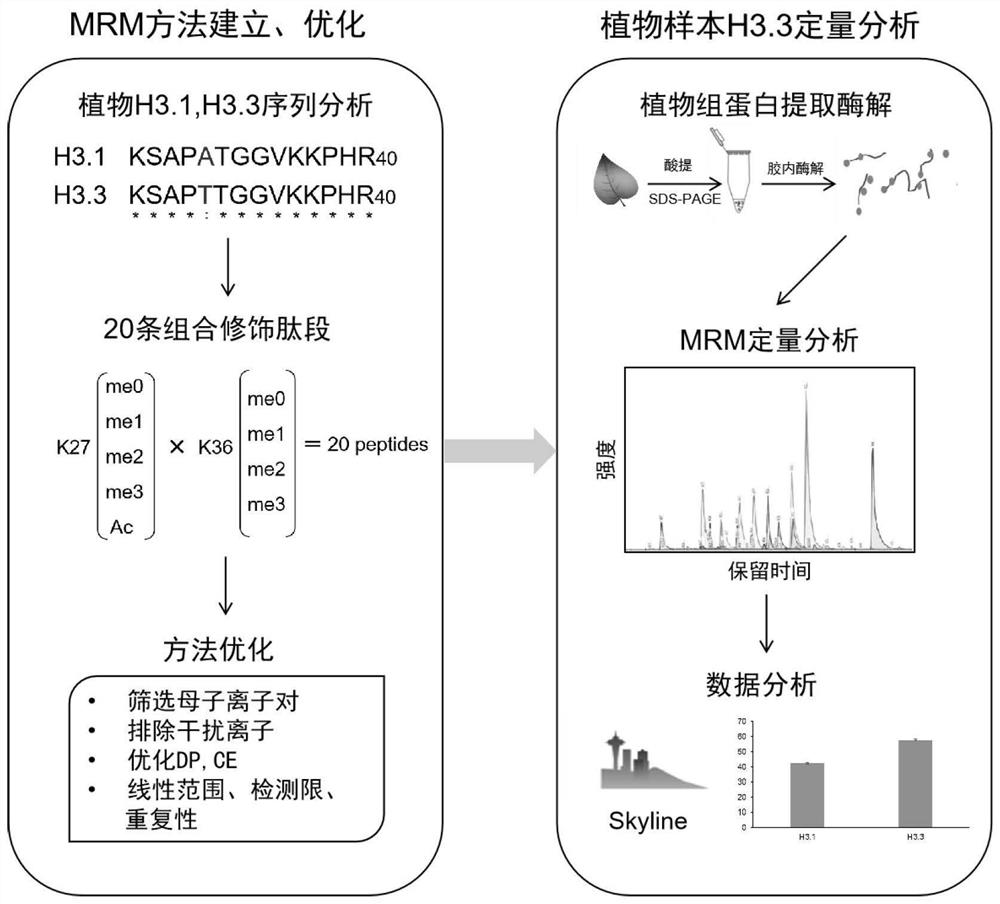
[0036] Example 1 Establishment and optimization of MRM-based quantitative detection method for plant histone variant H3.3 K27 / K36
[0037] 1. Synthesis of Plant Histone H3.3
[0038] The biological company synthesized the following sequence:
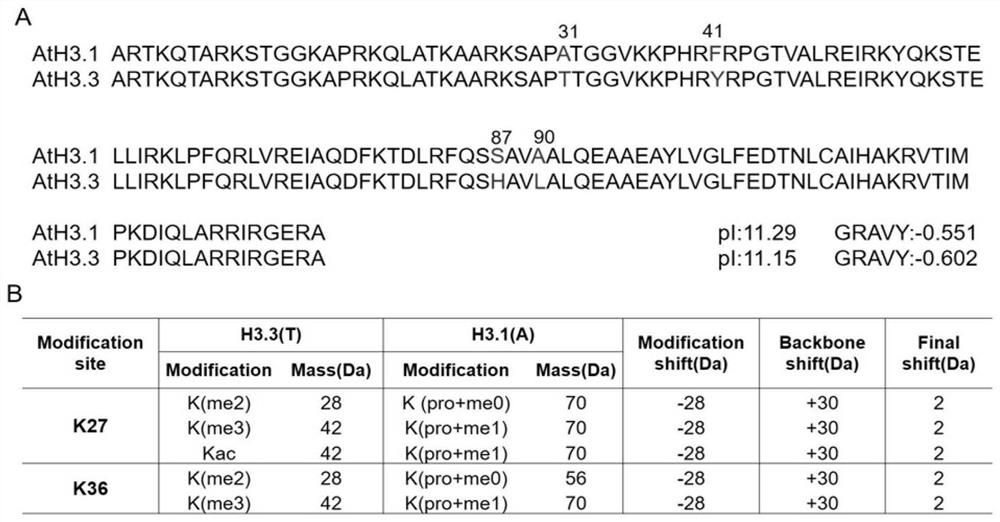
[0039] (ARTKQTARKSTGGKAPRKQLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA) K27-R40 peptides of 20 different combinations were dissolved in ddH 2 In O, until the final concentration is 100 μM, it is the mother (reservoir) solution;
[0040] 2. Dilute 1 μL of the mother solution to 100nM with 0.1% formic acid, enter the mass spectrometry, analyze the fragment ion response of each peptide, and select 3-5 product ions. The parent ion selection is good responsivity and has 3+ or 4+ charges, and the product ion selection is: 1) good responsivity; 2) product ion contains modification sites (K27 and K36); 3) between variants Product ions do not interfere.
[0041] 3. Optim...
Embodiment 2
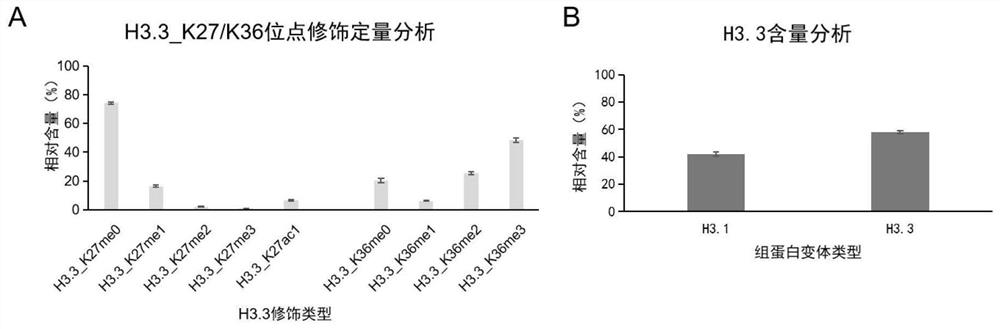
[0049] Example 2 Quantitative detection and analysis of histone variant H3.3 K27 / K36 in wild-type Arabidopsis
[0050] 1. Collect Arabidopsis thaliana seedlings about 15 days old, and extract core histones by acid extraction;
[0051] 2. Perform SDS-PAGE gel electrophoresis on the extracted crude histone, in-gel propionylation derivatization and enzymatic hydrolysis, as follows:
[0052] 1) Perform SDS-PAGE gel electrophoresis (12.5% or 15% gel) on the extracted crude histone, stain the protein with Coomassie Brilliant Blue overnight, and then decolorize until the background is almost transparent and colorless, and the bands are clear;
[0053] 2) Cut off the 15KD protein, mainly H3. Cut the strips into cubes as small as possible (1mm 3 left and right), placed in a centrifuge tube;
[0054] 3) Use about 500μL of ddH 2 O Low-speed shaking and washing for 15 minutes, repeat twice;
[0055] 4) Add 500 μL of decolorization solution (25mM NH 4 HCO 3 / 50%ACN) shake at room ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com