KOD DNA polymerase mutant, and preparation method and application thereof
A mutant and polymerase technology, applied in the field of KOD DNA polymerase mutants and their preparation, can solve the problems of inability to amplify gene fragments, reduced extension performance, etc., and achieve the expansion of application scope, improvement of inhibitor resistance, and PCR reaction performance. boosted effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Embodiment 1 constructs the recombinant vector containing the nucleotide sequence of coding KOFU-S DNA polymerase
[0058] (1) According to the amino acid sequence (SEQ ID NO.1) of the mutant KOFU-SDNA polymerase, after codon optimization of the E. coli expression system, a DNA molecule capable of high-efficiency expression in E. coli was obtained, and splicing was used to In the PCR method, the DNA molecule encoding the mutant KOFU-SDNA polymerase is artificially synthesized, specifically as shown in SEQ ID NO:2.
[0059] (2) The DNA molecule encoding the mutant KOFU-SDNA polymerase was recombined with the expression vector pET-28a. The synthetic primer sequences are as follows:
[0060] NdeI-FP: 5'-GCGC CATATG ATGTTAACCATC-3' (SEQ ID NO: 4);
[0061] XhoI-RP: 5'-GCGC CTCGAG TTATTTTTTCTG-3' (SEQ ID NO: 5).
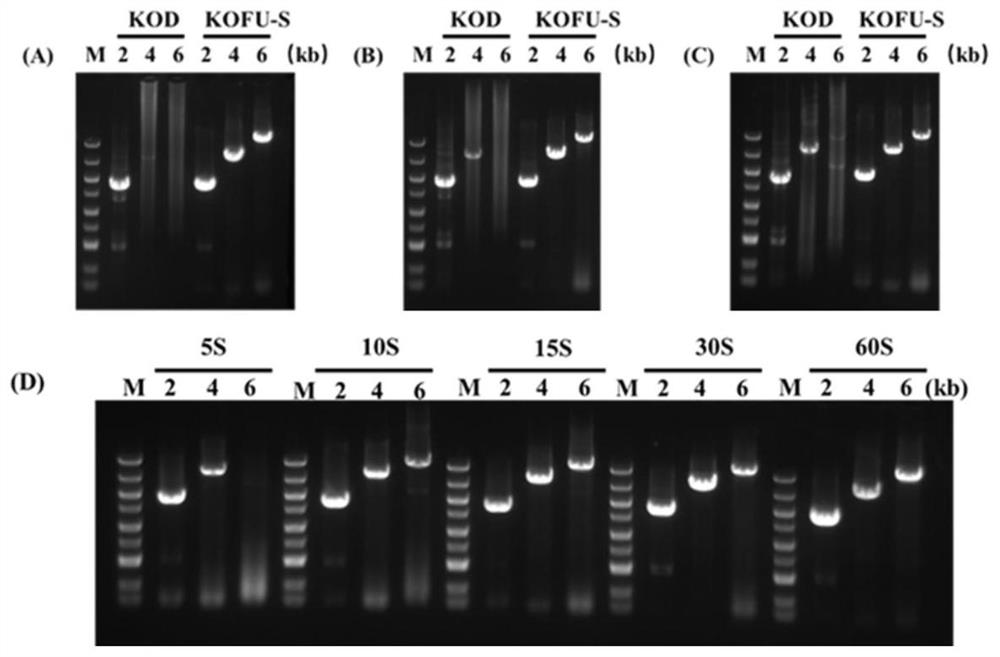
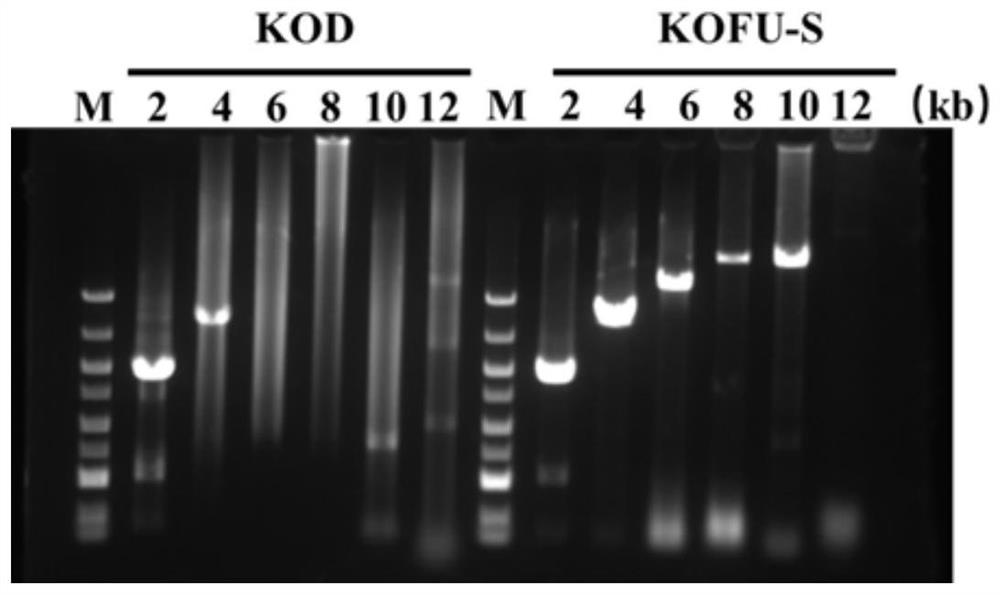
[0062] PCR was used to amplify the DNA molecule of the mutant KOFU-S DNA polymerase, and the artificially synthesized DNA molecule encoding the mutant KOFU-...
Embodiment 2
[0064] The preparation of the transformant of embodiment 2 expression mutant KOFU-SDNA polymerase
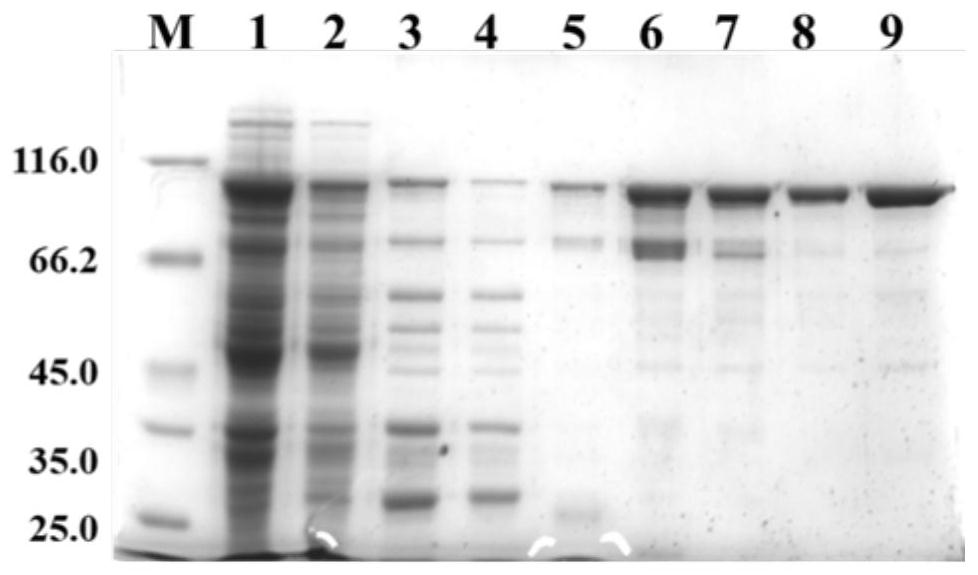
[0065] Transform the recombinant vector obtained in Example 1 into the host cell E.coli BL21(DE3), pick a single colony, inoculate into liquid LB medium and cultivate to OD 600 was 0.8, added IPTG to a final concentration of 0.1mmol / L, induced at 18°C for 16h, collected the cells and sonicated them, and then detected the expression of the target protein by SDS-PAGE electrophoresis. It was found that the prepared transformants could highly express the mutant KOFU- SDNA polymerase.
Embodiment 3
[0066] Expression and identification of embodiment 3 mutant KOFU-SDNA polymerase
[0067] The positive transformant strains capable of expressing the mutant KOFU-SDNA polymerase obtained in Example 2 were inoculated into 60 mL LB medium containing 50 μg / mL kanamycin sulfate, and placed in a shaker at 37° C. for shaking culture overnight; Take the seed solution cultivated overnight, inoculate it into 500 mL LB medium containing 50 μg / mL kanamycin sulfate at a ratio of 1:100, and continue shaking culture in a shaker at 37 ° C for 4 h (OD 600 =0.6~0.8); IPTG was added to each bottle of bacterial solution to a final concentration of 0.1mmol / L, and the shaking induction was continued at 18°C for 18 hours; the induced bacterial cells were collected by centrifugation and weighed, and the wet weight of the bacterial cells was recorded; 1 mL of each bacterial solution was centrifuged at 12,000 rpm for 1 min, and the precipitate was collected, and 150 μL of lysis buffer was added, osc...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com