Method of introducing target-specific foreign gene
An exogenous, genetic technology, applied to other methods of inserting foreign genetic material, cells modified by introducing foreign genetic material, using a vector to introduce foreign genetic material, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
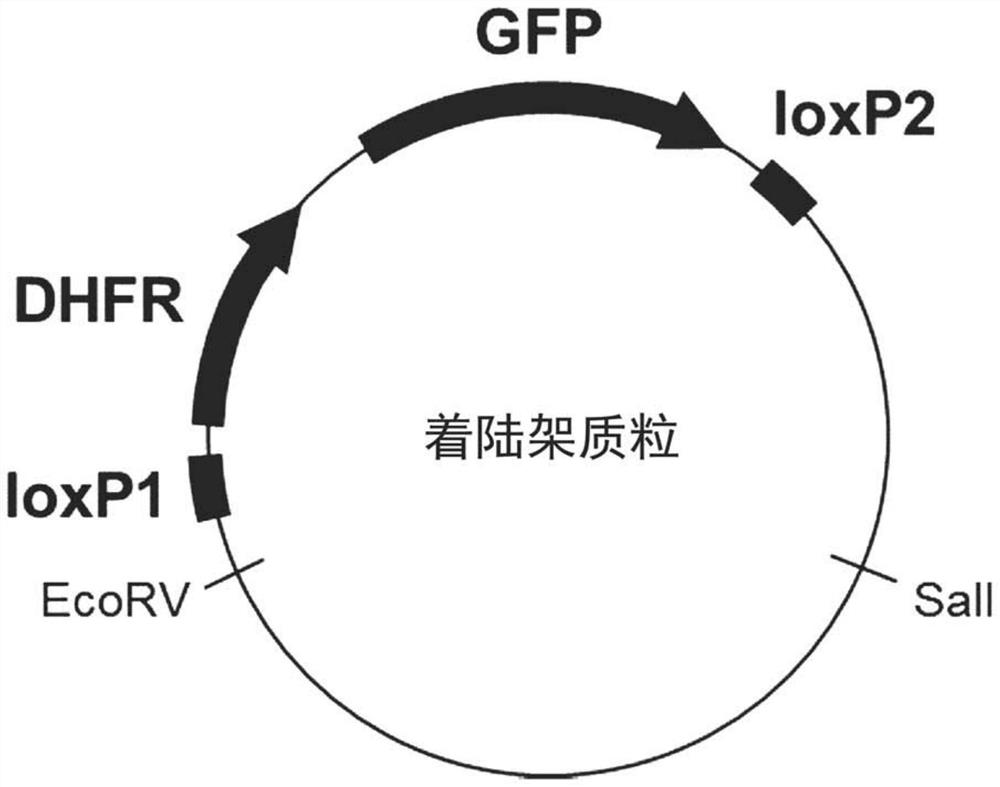
[0142] [Example 1] Preparation of Landing Pad Plasmid
[0143] Created a "landing rack" containing a DNA cassette that was used as a target site for the introduction of the gene of interest during the cassette exchange reaction and integrated it into the plasmid ( figure 1 ). The landing rack plasmid carries the green fluorescent protein (GFP) gene as a marker gene, which is used to identify gene high expression regions on the genome of CHO cells. It also has the dihydrofolate reductase (DHFR) gene as a selectable marker after the introduction of the landing shelf plasmid. DNA sequences (loxP1 and loxP2) recognized by recombinase Cre are inserted into both ends of the two genes, DHFR gene and GFP gene. The DHFR gene and the GFP gene present between these two loxPs were removed during the cassette exchange reaction and replaced with genes encoding the polypeptide to be produced loaded on the recombinant plasmid. In the following examples, an attempt was made to generate anti...
Embodiment 2
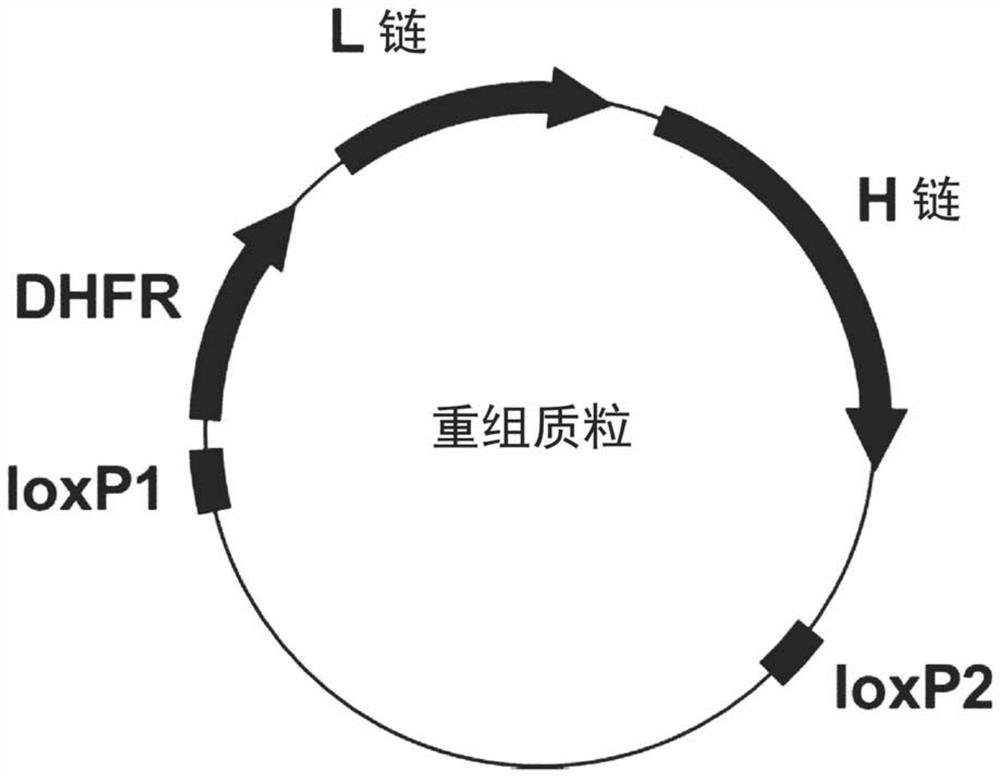
[0144] [Example 2] Preparation of recombinant plasmid
[0145] A "recombinant plasmid" was prepared for a cassette exchange reaction with the DNA cassette inserted into the landing pad on the CHO cell genome ( figure 2 ). The recombinant plasmid carries the DHFR gene and the antibody gene consisting of heavy and light chains, and loxP is inserted at both ends of these genes. As evaluation antibodies, one type of IgG1 antibody (mAb-A) and two types of IgG2 antibodies (mAb-B, C) were used.
[0146] Each antibody recognizes a different antigen as follows:
[0147] mAb-A: GYM329 / anti-latent myostatin antibody / IgG1;
[0148] mAb-B: CIM331 / anti-IL-31 receptor antibody / IgG2 (WO 2010 / 064697);
[0149] mAb-C: SA237 / anti-IL-6 receptor antibody / IgG2 (WO 2016 / 027859).
[0150] Although figure 2 A recombinant plasmid carrying antibody genes consisting of one set of heavy chain / light chains was used, but the configuration of the recombinant plasmid can be appropriately modified acco...
Embodiment 3
[0151] [Example 3] Preparation of host cells for site-specific integration (TI)
[0152] The landing pad plasmid was transfected into host cells (CHO-DXB11 ) using Nucleofector 2b from LONZA (Nucleofector is a registered trademark of Lonza Cologne GmbH). The landing pad plasmid used for transfection was linearized with the restriction enzymes EcoRV and SalI. Four hours after transfection, the medium was changed to a hypoxanthine / thymidine-free medium, and shaking culture of the cells was started. After about two weeks, single cell sorting was performed using Sony's Cell Sorter SH800. When sorting, sort the cell population within the top 2% with high GFP fluorescence intensity. A 488nm semiconductor laser was used to excite GFP. Single-cell sorted cells are expanded and cultured, and genomic DNA is extracted from each cell clone. Using the recovered genomic DNA, the copy number of the GFP gene introduced into each cell clone was measured using Bio-Rad's QX200 Droplet Digita...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com