Method for quantitative correction of mRNA detection in exosome
An exosome, detection value technology, applied in quantitative correction of fluorescent PCR. It can solve the problems of poor stability and inability to distinguish tumor patients from healthy people, and achieve the effect of improving the degree of discrimination, sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Screening of urine exosome internal reference genes
[0034] 1.1 Urine exosome preparation and exosome RNA extraction
[0035] 1) Urine sample preparation, the required amount is 30mL. Take 9 urine samples from healthy volunteers, numbered N1-N9; 9 urine samples from urothelial cancer patients, numbered T1-T9;
[0036] 2) Take a urine sample, centrifuge at 3000g at 4°C for 15min, transfer the supernatant to a new centrifuge tube, add 15mL of PEG8000 aqueous solution (concentration 30%), shake and mix, let stand overnight at 4°C, then, 3000g, Centrifuge for 10 min, and use the Qiagen miRNeasy Micro Kit (Qiagen, 217084) to extract exosome RNA from the obtained pellet;
[0037] 1.2 Urine exosomal RNA library construction
[0038] The urine exosome library was constructed using the Ovation SoLo RNA-Seq System kit (NuGen, catalog number: M01406). For the operation method, please refer to the kit instruction manual.
[0039] 1.3 Send the urine exosomal RNA librar...
Embodiment 2
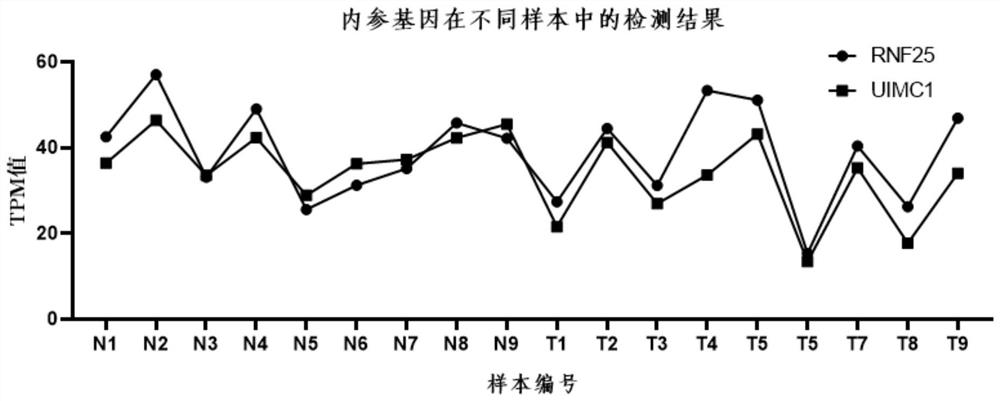
[0049] Example 2 The differential genes screened in urine exosome sequencing samples were corrected using the screened internal reference gene RNF25 / UIMC1
[0050] Through calculation and analysis, we found that for the internal reference gene RNF25, the maximum detection value among the 18 samples was 57.02, and the minimum detection value was 15.31, with a difference of 3.72 times; for UIMC1, the maximum detection value among the 18 samples was 46.36, the minimum detection value The detection value is 13.4, the difference is 3.45 times, and the relative expression of the two genes (RNF25 / UIMC1) trend is between 0.86-1.59 (see Table 3), this fluctuation is relatively small, as shown in Table 3 Show.
[0051] Table 3. Calculation of expression difference and relative expression of internal reference genes
[0052] gene_name N1 N2 N3 N4 N5 N6 N7 N8 N9 RNF25 42.5 57.02 33.15 49 25.57 31.2 35.1 45.76 42.18 UIMC1 36.39 46.36 33.53 42.32 ...
Embodiment 3
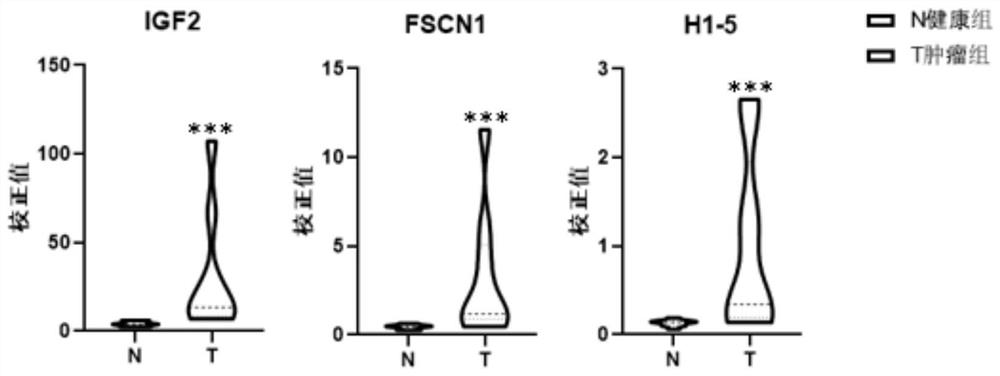
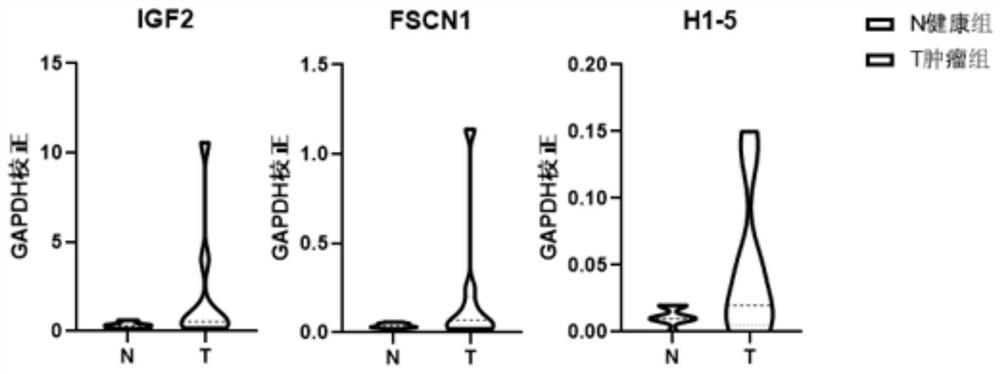
[0063] Example 3 qPCR verification of urinary exosome RNF25 gene and UIMC1 gene among different people
[0064] In order to test the relative stability of the RNF25 gene and UIMC1 gene in different samples, we collected 7 urine samples from healthy people, numbered N1-N7, and prepared exosomes and exosomal RNA. The preparation of exosomes and exosomal RNA refers to the process shown in Example 1. After that, the two genes were verified by qPCR. The sequence information of the two primers and probes is shown in Table 7. The qPCR detection uses the Fapon one-step reagent , One step RT-PCR kit (MD027, Faipeng Biotechnology Co., Ltd.), the configuration of the sample well reaction system is as follows:
[0065] Element Volume (μL) RT-PCR Master Mix 10 Enzyme Mix 3 Primer Probe Solution 2 template 5 Enzyme-free water 30
[0066] The reaction process is as follows:
[0067]
[0068] Table 7. RNF25 gene and UIMC1 gene primer and prob...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com