Preparation method of protein containing non-natural amino acid
An unnatural amino acid and aminoacyl technology, applied in the field of biopharmaceuticals, can solve problems such as the lack of research methods for small molecule drug systems, the reduction of drug efficacy caused by neutralizing antibodies, and the modification of multiple proteins with different unnatural amino acids
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0086] Example 1 Four kinds of unnatural amino acid systems expanded into 12 different systems
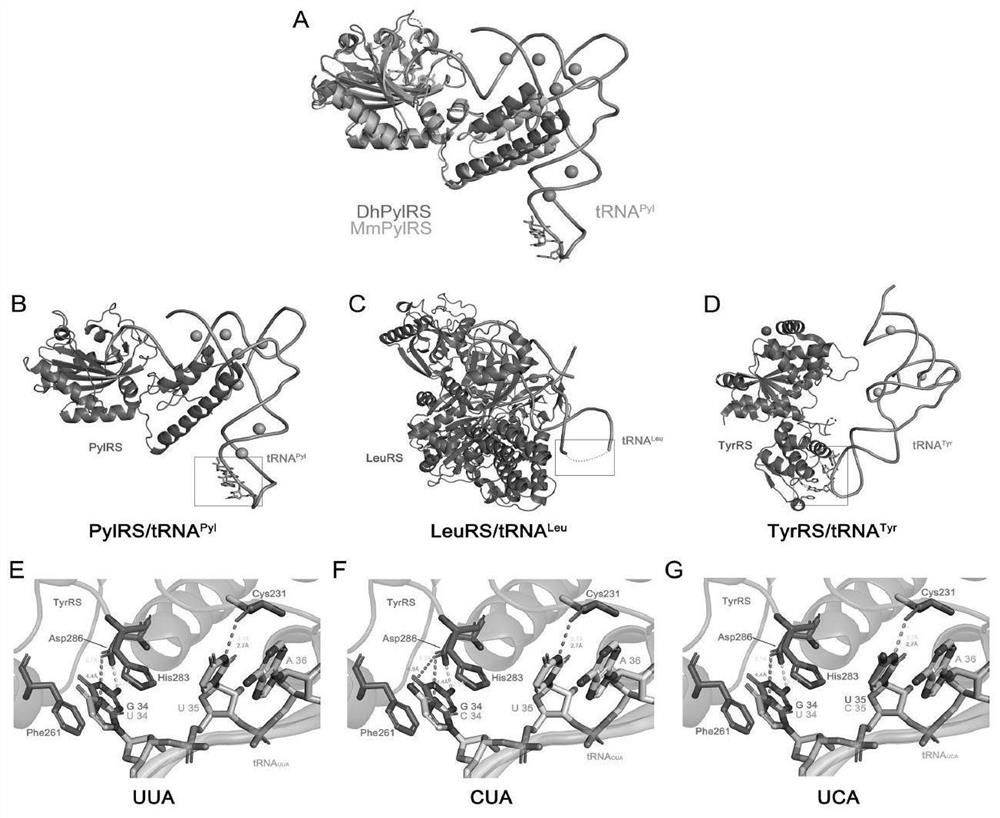
[0087] We used available complexes of Desulfobacillus hafniense except for pylMmRS (PDB:2ZIM), two synthetases that are structurally conserved. According to the binding structure of synthetase and corresponding tRNA, tRNA Pyl and tRNA Leu The anticodon loop of is not bound by its synthetase. The anticodon loop of tRNATyr does form hydrogen bonds with D286 and C231 in TyrRS, but the mutated anticodons UUA, CUA, and UCA maintain weak interactions with the aforementioned key residues in TyrRS, thus ensuring successful aminoacylation (see figure 1 ).
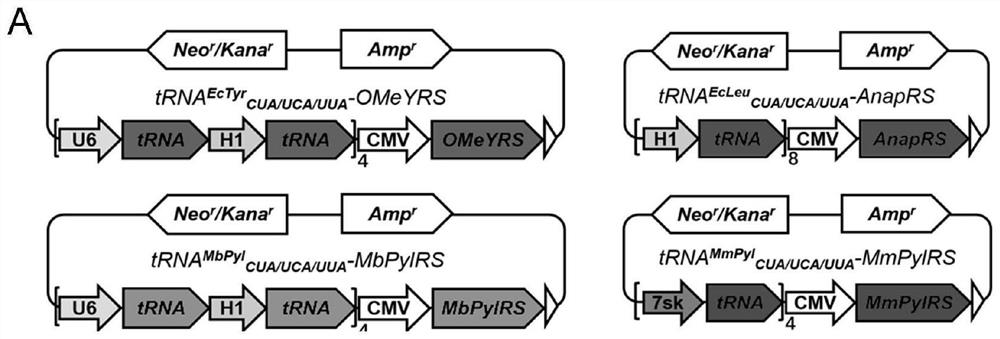
[0088] In order to construct plasmids encoding four aminoacyl tRNA synthetases (OMeYRS, EcleuRS, MbPylRS and MmpylRS), the corresponding genes expressed under the CMV promoter were inserted into the pcDNA3.1(+) vector by double digestion reaction. tRNA MbPyl and tRNA EcTyr Expressed by H1 and U6 promoters, tRNA Ecleu Expressed by ...
Embodiment 2
[0095] Example 2 Orthogonality verification of four unnatural amino acid systems and screening of the best combination of the three systems with the highest efficiency in mammalian cells
[0096] To rule out interactions of different systems, we verified the mutual orthogonality of the four available aaRS / tRNA systems ( Figure 3C ).
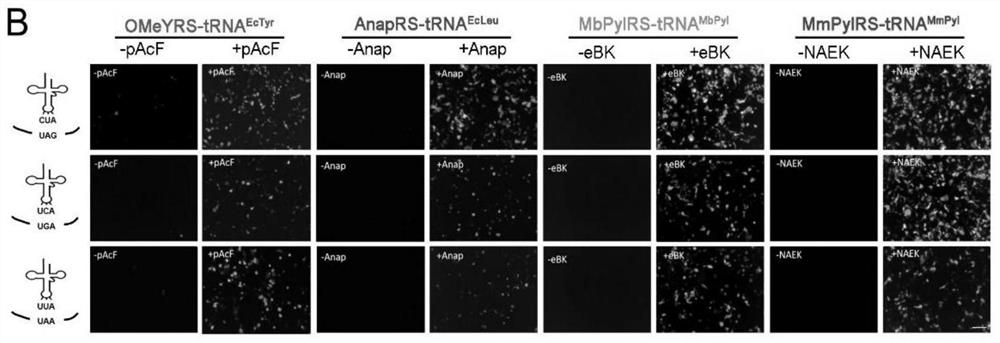
[0097] First, we performed orthogonality verification at the UAA and aaRS / tRNA pair levels. 48 hours after the transfection system and the corresponding GFP plasmid, different unnatural amino acids were added to the culture medium, the fluorescence of GFP was observed, and the fluorescence intensity was compared by flow cytometric analysis. The results are as follows Figure 3A shown.
[0098] We then determined orthogonality at the level of synthetases and tRNAs. Synthetases were co-transfected with different tRNAs CUA In the HEK293T cells, as well as the GFP containing the TAG mutation, observe the fluorescence of GFP, and perform flow cyt...
Embodiment 3
[0106] Example 3 Plasmid construction of RFP, BFP, GFP and controllable expression of three fluorescent proteins in mammalian cells
[0107] We first constructed the RFP based on point mutation and molecular cloning methods 36TGA 、BFP 39TAG and GFP 39TAA Mutants, primers for constructing the above three mutant fluorescent proteins are shown in Table 5.
[0108] Table 5. Primer sequences used in the construction of three fluorescent protein mutants
[0109] Primer sequence GFP 39TAA -F
[0110] System plasmid containing Mmpyl-tRNA pyl UCA ,EcTyr-tRNA Tyr UUA , Ecleu-tRNA leu CUA , transfected them together into HEK293T cells, designed 8 groups of experiments, added 0, 1, 2, and 3 different unnatural amino acids to the medium, collected the cells after 48 hours, and extracted the protein for Western blotting. The expression of three fluorescent proteins was detected under different experimental conditions. For specific results, see Figure 4 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com