Imaging-based pooled crispr screening
A reporter gene and sequence technology, applied in the field of cell imaging, can solve problems such as difficulty in determining the genotype of a single cell
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
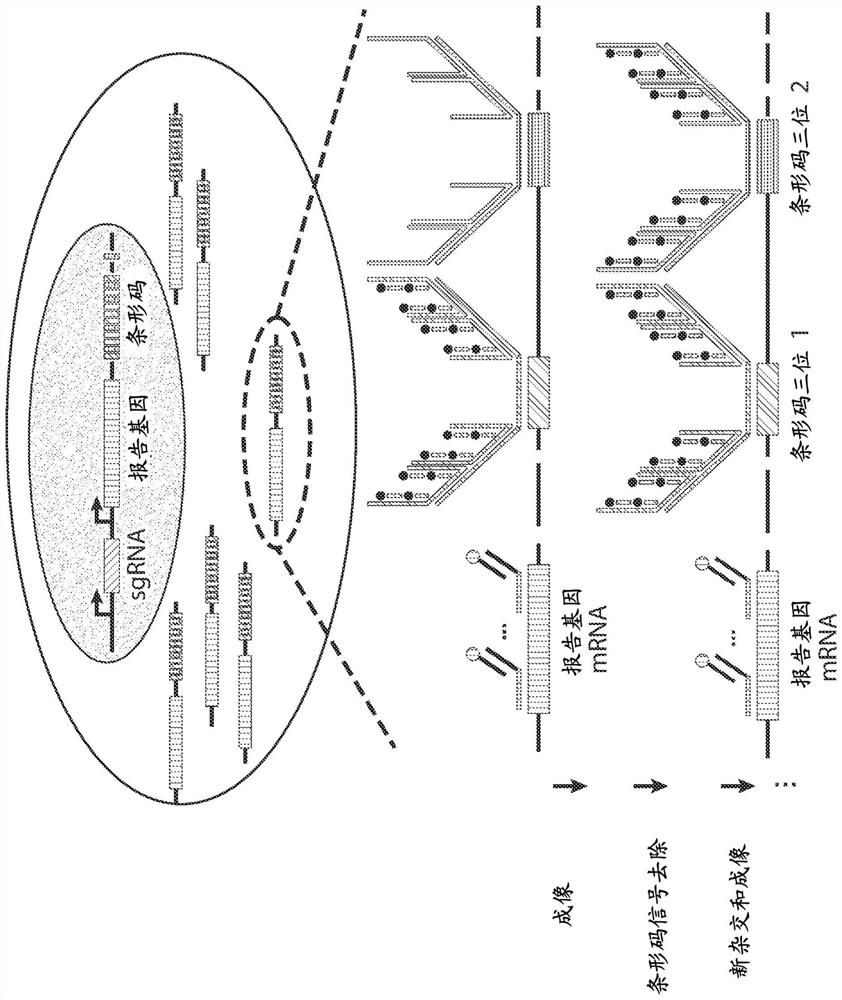
[0177]CRISPR screening of pooled libraries provides a powerful means to discover genetic factors involved in cellular processes in a high-throughput manner. However, the phenotypes available for pooled library screening are limited. Complex phenotypes, such as cellular morphology and subcellular molecular organization, as well as their dynamics, require imaging-based readouts that cannot currently be derived using pooled library CRISPR screens. These examples demonstrate a fully imaging-based hybrid library CRISPR screening approach that combines high-content phenotypic imaging with high-throughput guide RNA (sgRNA) identification in single cells. In one such approach, sgRNAs are co-delivered to cells with corresponding barcodes placed in the 3' untranslated region (3'UTR) of the reporter gene using a lentiviral delivery system with reduced recombination Induced sgRNA-barcode mismatches. Barcodes can be read out using multiplex error robust fluorescence in situ hybridization...
Embodiment 2
[0181] This example illustrates high-throughput high-precision barcode imaging in mammalian cells. In situ imaging-based screening of pooled libraries has been performed in bacteria in recent years, where the genotype of individual cells is identified by multiplex FISH imaging of barcodes associated with gene variants. Because of the small size of bacterial cells, the diffuse signal from barcoded RNA in a single cell is strong enough to be easily measured. However, mammalian cells are approximately thousand times larger than bacteria, making it difficult to achieve high enough concentrations of barcoded RNA to allow reliable measurements. Therefore new barcode expression and detection protocols are needed for mammalian cells to increase barcode signal and reduce background.
[0182] To achieve this goal, two independent promoters were used to express the sgRNA and the reporter gene in the same vector and a 12-digit ternary barcode ( Figure 1A ). Each digit (digit) of the t...
Embodiment 3
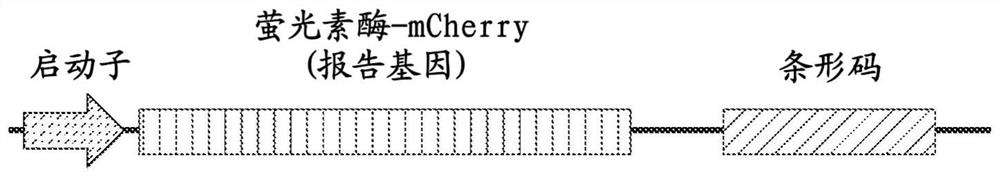
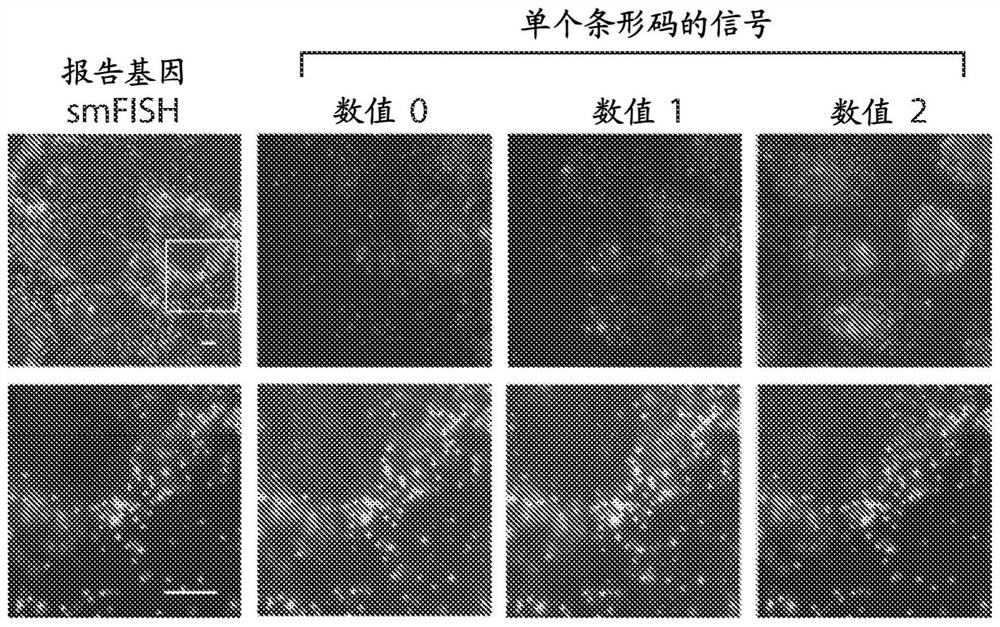
[0190] To further verify the low mislabeling rate, in this example, two reporter gene-barcode libraries were designed, each expressing a reporter with a different epitope tag (HA tag or Myc tag) fused to the barcode library as described above. gene luciferase-mCherry ( Figure 2A ), and the two libraries were cloned separately. Each library was narrowed down to contain Figure 2B ), the barcodes associated with each cell were imaged using successive rounds of hybridization as described above. The rationale is that determining the phenotype of each cell will allow the barcode identity of that cell to be deduced from the sequencing results, and then comparison with barcodes determined by imaging will allow the determination of a small fraction of misidentified barcodes.
[0191] Judging by barcode-phenotype mismatches, only ~1% of cells had incorrectly identified barcodes ( Figure 2C and 2D ). Even this small error is likely due to errors in cell segmentation, which in turn...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com