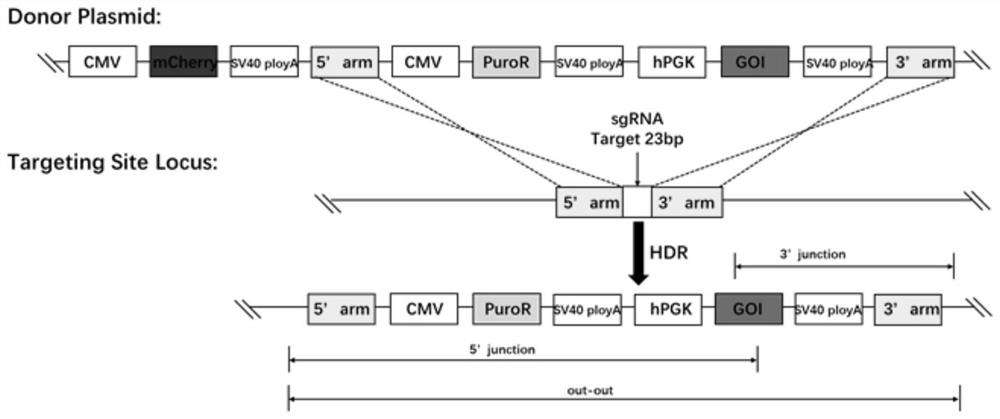
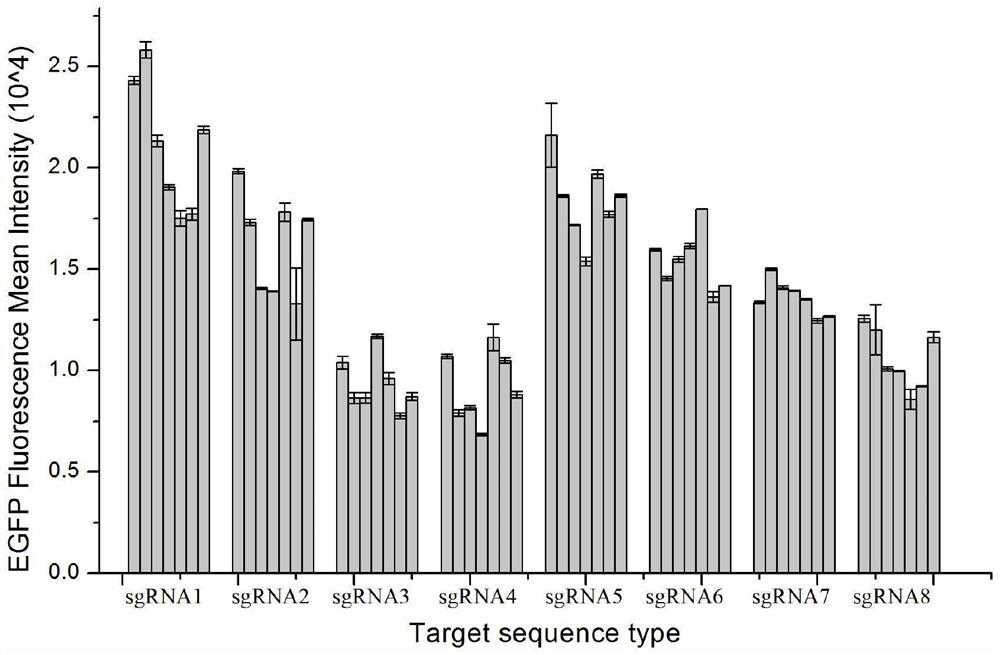
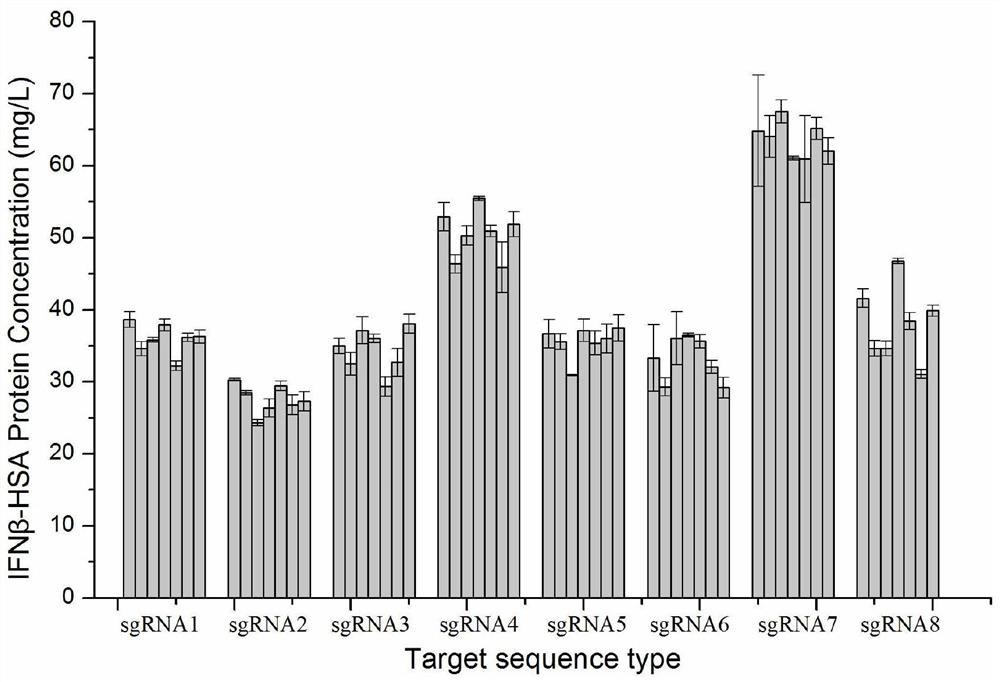
Site for stably expressing protein in CHO cell gene NW_003613781.1 and application of site
A stable expression and cell gene technology, applied in the field of genes, can solve the problems of long time consumption, stability affecting product launch time, instability of recombinant CHO cell lines, etc., and achieve the effect of reducing costs and shortening research and development time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1: Screening of stable expression sites
[0039] The CHO-K1-1b7 cells expressing the Zsgreen1 reporter gene screened by high-throughput flow cytometry were cultured in an adherent culture until they were in good condition, and the CHO cells at this time were regarded as the 0th generation, and then cultured continuously for 20 The expression of Zsgreen1 protein in cells at passages 0, 10, and 20 was observed under an inverted fluorescence microscope, and the average fluorescence intensity of cells at passages 0, 10, and 20 was detected by BD flow cytometry.
[0040] Observation by inverted fluorescence microscope and detection by flow cytometry show that after 20 passages, CHO-K1-1b7 cells can still express 100% Zsgreen1 protein in the adherent culture state, and Zsgreen1 protein between different passages The expression levels are basically the same, and there is a strong green fluorescent signal.
[0041] The CHO-K1-1b7 cells that passed the verification of a...
Embodiment 2
[0043] Example 2: Analysis of Lentiviral Integration Sites
[0044] Use the Lenti-X Integration Site Analysis Kit (Clontech: 631263) related to chromosome walking technology to analyze the integration site of the lentiviral vector in CHO-K1-1b7 cells. The specific steps are as follows:
[0045] (1), construction of lentiviral integration library
[0046] Collect CHO-K1-1b7 cells, use the DNA extraction kit to extract the genome, use DraI, SspI, and HpaI three restriction endonucleases to digest the genome at 37°C for 16-18h, and the digestion system is as follows:
[0047]
[0048] The digested product was purified and recovered using a PCR purification kit, and the chromosome walking adapter GenomeWalker Adapter was connected to both ends of the purified digested fragment. The connection system was as follows:
[0049]
[0050] Incubate overnight at 16°C and incubate at 70°C for 5 minutes. After the reaction is terminated, 32 μl TE (10 / 1, pH 7.5) is added to the system...
Embodiment 3
[0065] Example 3: Target Sequence Selection
[0066] 根据就近原则,使用CCTOP CRISPR / Cas9在线预测系统对位点NW_003613781.1第1497187碱基处上下游各100bp的序列:AAAATGTGTATAACTCCAGATTCAGAGGATCCAACACTTTCCACTGGCATGTGCATATAGTGCGCATACATTTGTATAGGCTAAAAACTCATAATATTATACAATAACAATAATAATGATAAATCTGTAATAACAATCTCTATCCATTATTCACTATTTCGGTATTTGCCAAACCAAACAAAATCCTTCCTATCTTTTCT(SEQ ID NO.1)进行预测,并挑选出编辑效率较高的靶 sequence.
[0067] The relevant parameters are set as follows:
[0068] 1) The maximum number of base mismatches allowed in the first 13 bp within the 20 bp sequence after NGG is 1;
[0069] 2) The number of all 20bp mismatched bases after NGG is 4.
[0070] The CCTOP CRISPR / Cas9 online prediction system will score the editing efficiency of the identified 5'NNNNNNNNNNNNNNNNNNNNNGG 3' target sequence, LOW efficiency(score0.74 ).
[0071] Sequences in which the predicted editing efficiency was higher than 0.56 were selected as target sequences.
[0072] Target sequence 5'-GAGGATCCAACACTTTCCACTGG-3'(SEQ ID NO.2), score=0.68 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com