Detection method and kit for diagnosing bladder urothelium carcinoma
A technology of urothelial carcinoma and a kit, which is applied in the fields of biotechnology and medical diagnosis, can solve the problems of low sensitivity and specificity, unobvious early symptoms of bladder urothelial carcinoma, and difficulty in detection, and achieve excellent sensitivity and specificity sexual effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] The present invention is based on performing single-cell transcriptome, single-cell chromatin accessibility sequencing and Whole-genome DNA methylation sequencing, analysis found that cells of different origin types have different DNA methylation characteristics, causing differences in the degree of chromatin opening, which in turn affects differences in gene expression, and ultimately leads to different fates of cells. Some are normal cells, while some develop into muscle-invasive cancer cells, or non-muscle-invasive cancer cells. Through statistical analysis of these DNA methylation features, regions (DMRs) with significant differences in methylation levels in the three source cells were screened out, as shown in Table 1. The present invention is based on the method of methylation multiplex PCR, amplifies these DMR regions, and detects the methylation level on these DNA regions by high-throughput sequencing, and the specific primer combinations are shown in Table 2: ...
Embodiment 2
[0058] Seventeen methylated regions were detected using commercial fully methylated and unmethylated pattern standards (purchased from Zymo).
[0059] The specific process is as follows:
[0060] 1. DNA bisulfite conversion (BS treatment)
[0061] The DNA bisulfite conversion kit was purchased from Zymo, and was carried out according to the instructions of the kit, and the conversion product was obtained with NF-H 2 O was eluted for later use.
[0062] 2. Model Standard Gradient Incorporation
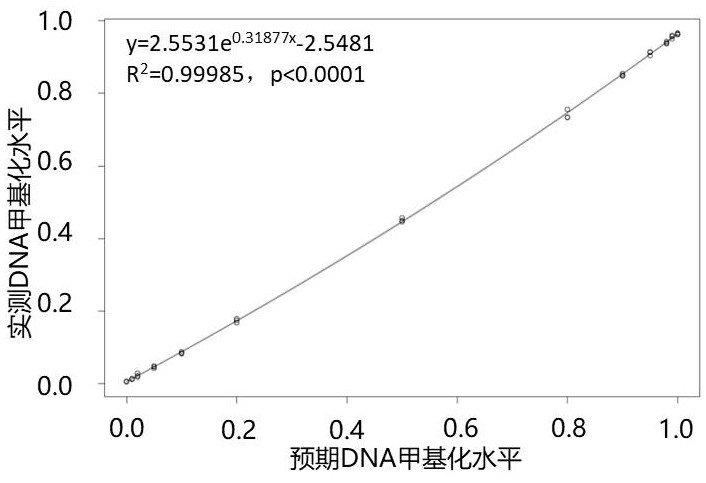
[0063] Single-strand quantification was performed after BS-treated fully methylated and non-methylated standard samples were disrupted by ultrasound. Gradient incorporation was carried out according to the DNA concentration, and 13 model standard gradients were set up in this application (methylation levels were 100%, 99%, 98%, 95%, 90%, 80%, 50%, 20%, 10% %, 5%, 2%, 1%, 0%).
[0064] 3. Preparation of Methylated Multiplex PCR Primer Sets
[0065] 19 pairs of primers were mixed fo...
Embodiment 3
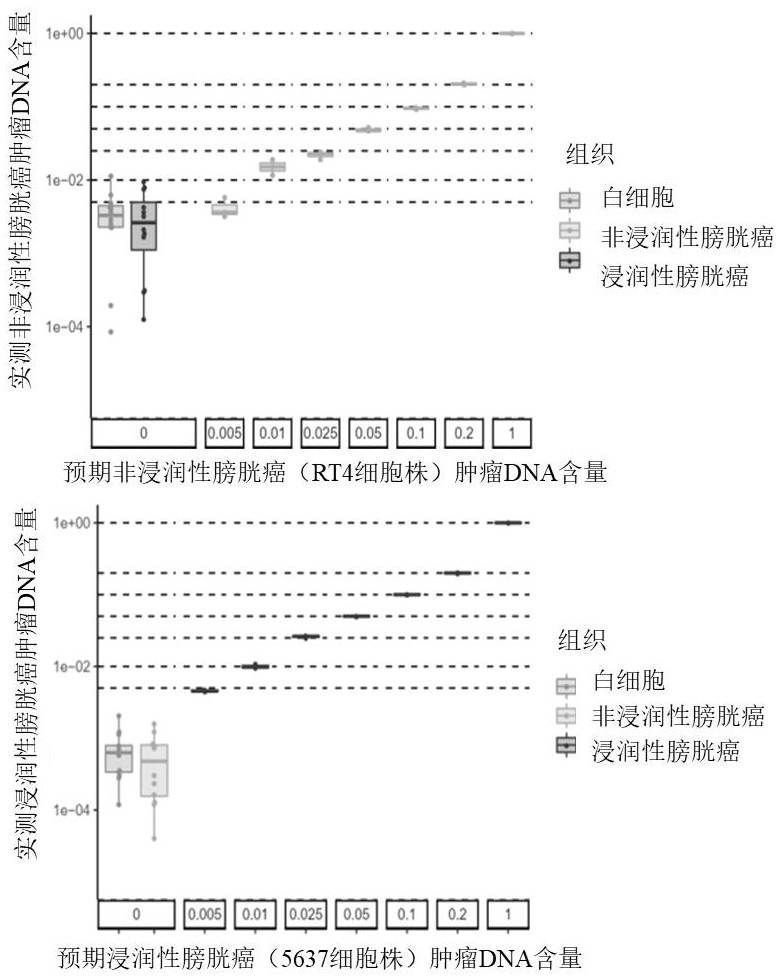
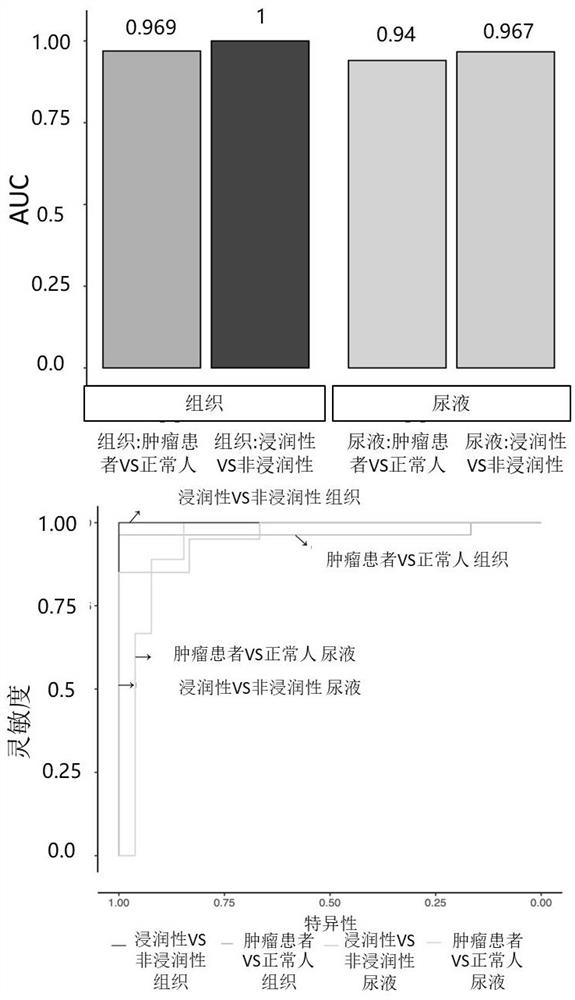
[0105] Use bladder urothelial cancer cell lines RT4 and 5637 to detect biological standards to clarify the minimum detection limit of this application for bladder urothelial cancer and its typing
[0106] The specific process is as follows:
[0107] 1. gDNA extraction
[0108] Bladder urothelial carcinoma cell lines were purchased from the Cell Bank of the Chinese Academy of Sciences. The extraction kit was purchased from QIAGEN (69506), and was carried out according to the instructions of the kit.
[0109] 2. DNA bisulfite conversion (reference example 2)
[0110] 3. Biological Standard Gradient Incorporation
[0111] The RT4 / 5637 gDNA treated with BS was ultrasonically fragmented to simulate cfDNA, and normal urine cfDNA did not need ultrasonic fragmentation to perform single-strand quantification. Gradient incorporation is carried out according to the DNA concentration. In this application, a total of 8 model standard gradients are set up for each group (the gDNA conten...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com