Genetically engineered bacterium for producing free fatty acid under assistance of formate
A technology of free fatty acid and genetically engineered bacteria, applied in the field of genetically engineered bacteria, can solve the problems of high energy consumption and difficult to obtain precursors, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
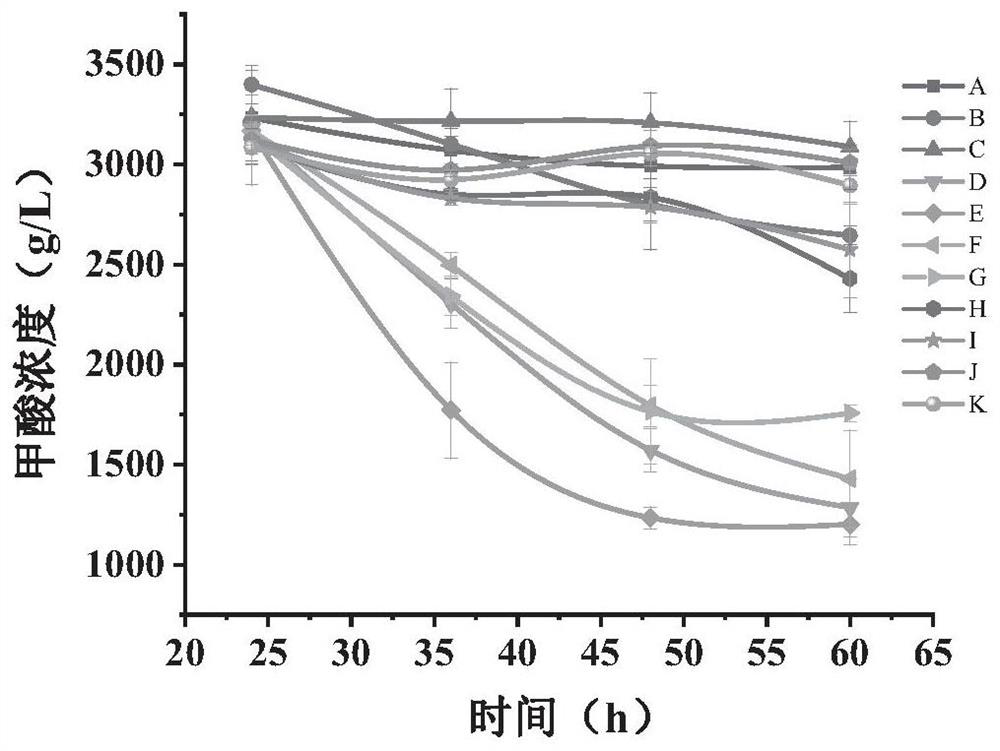
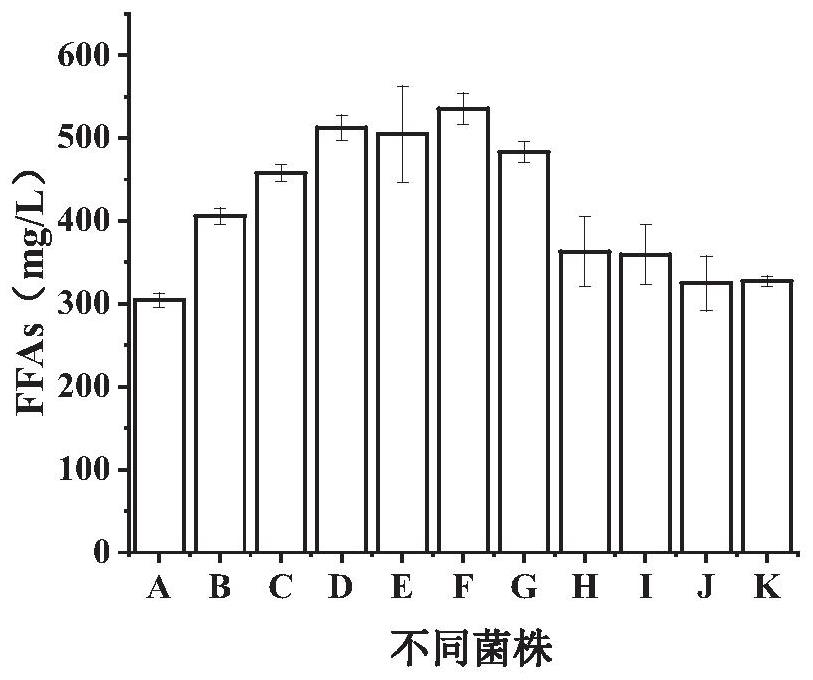
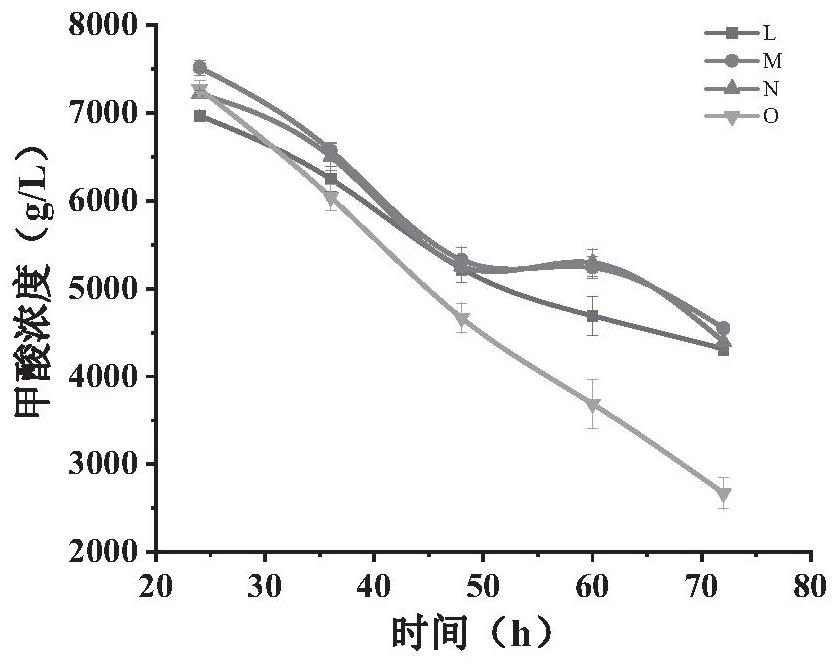
[0049] Existing microorganisms as hosts utilize formic acid and CO 2 Among the technologies, most of them use gene editing methods to modify the chassis of microorganisms so that they have this function, and to further improve the use of formic acid and CO by microorganisms 2 method and utilization of formic acid and CO 2 Proceeding to the production of microbial chemicals remains to be explored. In view of this, the present inventors use formic acid and CO 2 A certain amount of research has been carried out on the method of improving microbial fermentation to produce FFAs, a derivative product of acetyl-CoA. The specific research process is as follows:
[0050] The present invention selects Saccharomyces cerevisiae with mature gene manipulation technology and high robustness to perform formic acid and CO 2 utilization to produce FFAs. Formate can be used as an auxiliary carbon source, which can be converted into carbon dioxide under the action of formate dehydrogenase, ac...
Embodiment 1
[0131] Embodiment 1: Construction of knockout plasmid
[0132] The primers used in this example are shown in Table 1 above.
[0133] First, design the gRNA required for gene knockout through the website (https: / / www.atum.bio / eCommerce / cas9 / input), design the gRNA on the primer, and use the Pstg.Ura plasmid as a template to amplify the gene with uracil ( Ura) screened the marked fragments gRNA-hfd1, gRNA-pox1, gRNA-faa1, gRNA-faa4, and connected these four fragments with the plasmid pLacZ-SalI in turn, and sequenced to obtain the correct plasmids pCas9-hfd1, pCas9-pox1, pCas9 -faa1, pCas9-faa4. Subsequently, each gene was knocked out according to the traditional Cas9 knockout method of Saccharomyces cerevisiae gene.
Embodiment 2
[0134] Embodiment 2: Construction of recombinant plasmid
[0135] The primers used in this example are shown in Table 2 above.
[0136] The exogenous genes FDH1, FDH2, FDH3, FDH4, FDH4-TB1, FDH4-TB2, FDH4-TB3, FDH4-TB4, FDH4-TB5, FDH4-TB6 were synthesized by the company and connected to the plasmid Psp-GM1 respectively, After the experimental screening, the high-efficiency FDH was amplified using the respective plasmids as templates, and then integrated into the Psp-GM1 plasmid by restriction enzyme digestion, and the correct plasmids Psp-GM1-FDH3-FDH4 and Psp-GM1-FDH3 were obtained by sequencing - FDH4-TB1, Psp-GM1-FDH4-FDH4-TB1, Psp-GM1-FDH3-FDH4-FDH4-TB1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com