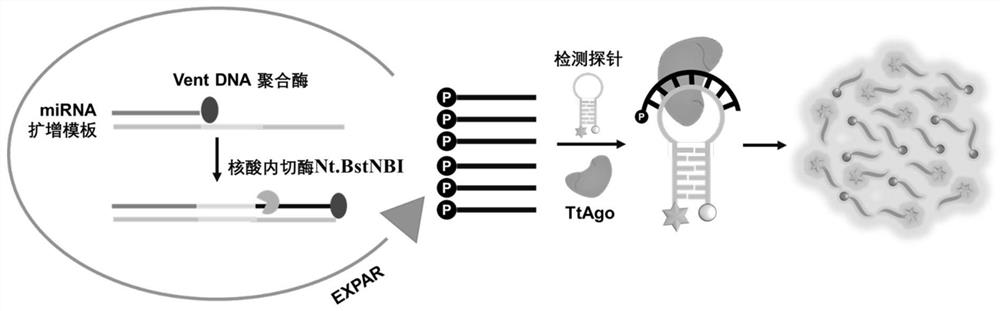
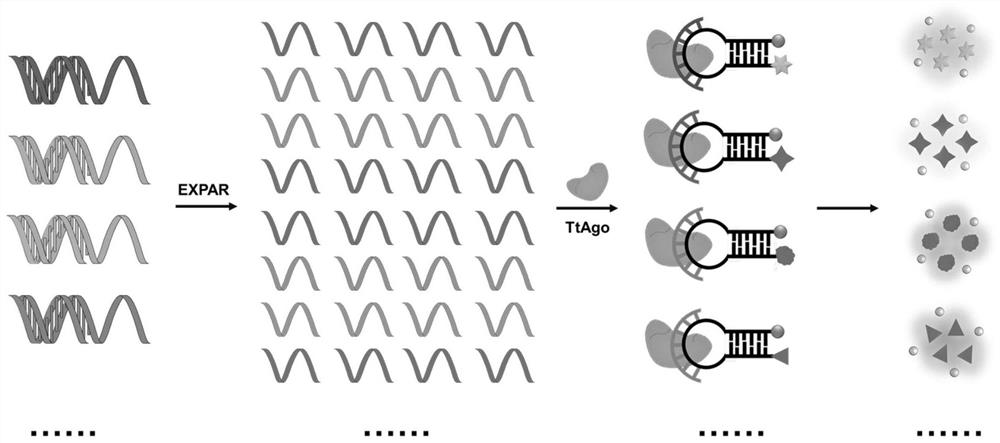
MicroRNA detection system and method based on exponential amplification reaction and Argonaute nuclease
An exponential amplification and reaction system technology, applied in the field of microRNA detection, can solve the problems of serious false positives, low abundance, and low detection efficiency, and achieve the effect of improving diagnostic accuracy and sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0107] Example 1: High specificity detection capability of TtAgo single base resolution
[0108] This example explores the cleavage specificity of TtAgo. Firstly, miDNA-21 and single base mismatched miDNA-21 (as shown in Table 2), and mock probes (ie, detection probes), as shown in Table 3, were synthesized.
[0109] Among them, miDNA-21 is 100% complementary to the simulated probe, and there is a single base mismatch between miDNA-21 and the simulated probe. Use miDNA-21 to simulate the EXPAR pre-amplification product of miRNA-21, the detection system based on TtAgo nuclease contains miDNA-21 (5μM) or single base mismatched miDNA-21 (5μM), mock probe (1μM) , TtAgo (100nM), Reaction buffer, MnCl 2 (750μM) and ultrapure water. The reaction temperature of the detection system based on TtAgo nuclease is 80° C.; the reaction time is 30 minutes. In the detection system based on TtAgo nuclease, the cleavage product (short-chain nucleic acid) of TtAgo nuclease is verified by 15...
Embodiment 2
[0113] Embodiment 2: the sensitivity of the miRNA detection method based on EXPAR and TtAgo system
[0114] This example explores the sensitivity of the miRNA detection method based on EXPAR and TtAgo nuclease.
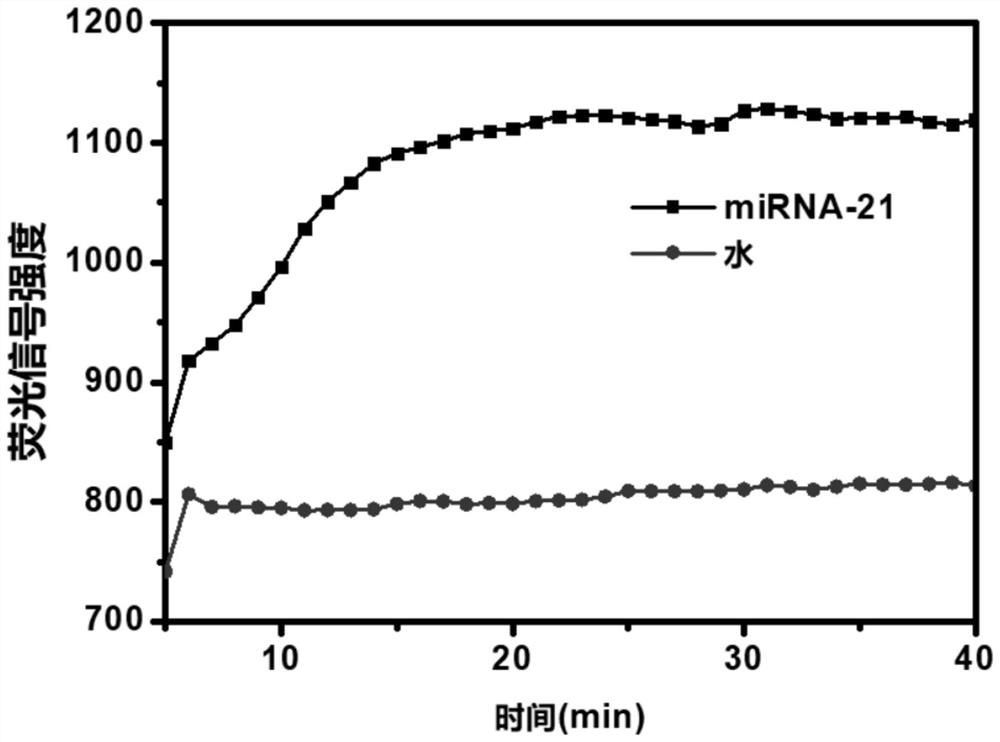
[0115] Taking miRNA-21 as an example, a series of concentrations of miRNA (as a nucleic acid sample) (0, 1aM, 10aM, 100aM, 1fM, 10fM, 100fM, 1pM, 10pM, 100pM, 1nM, 10nM) for pre-amplification, the reaction conditions are 55°C, 20min. The detection system based on TtAgo nuclease contains preamplification product (1μL), detection probe (1μM), TtAgo (100nM), Reaction buffer, MnCl 2 (750μM) and ultrapure water. The reaction temperature of the detection system based on TtAgo nuclease is 80° C.; the reaction time is 30 minutes. Test results such as Figure 5 As shown in A, the detection sensitivity reaches 1aM (10 -18 M)( Figure 5 in B).
[0116] In this example, the sensitivity of the miRNA detection method based on EXPAR and TtAgo nuclease was further compared w...
Embodiment 3
[0117] Embodiment 3: The miRNA detection method based on EXPAR and TtAgo nuclease detects multiple miRNA
[0118] This example explores the ability of the miRNA detection method based on EXPAR and TtAgo nuclease of the present invention to detect multiple miRNAs.
[0119] Taking the four target miRNAs of miRNA-21, miRNA-92a, miRNA-31 and miRNA-14 as examples, the detection process is divided into two steps, first based on the EXPAR pre-amplification system described in "Materials and Methods" , in the EXPAR pre-amplification system, add the amplification templates corresponding to four miRNAs and the corresponding four target miRNAs (as nucleic acid samples), the target miRNA concentration is 100pM, these four target miRNAs in the same reaction tube At the same time, pre-amplification was carried out, and the reaction conditions were 55° C. for 20 min. Secondly, the miRNA detection system based on TtAgo nuclease includes detection probes corresponding to four amplicons and EX...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com