Compositions and methods for neurological diseases
A technique of combining and combining structural domains, which can be applied to neurological diseases, chemical instruments and methods, receptors of neurotransmitters, etc., and can solve the problems that gene therapy has not been widely used to treat neurological diseases
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
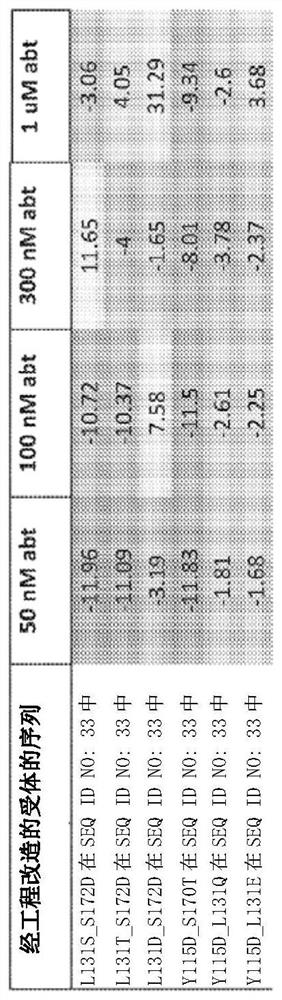
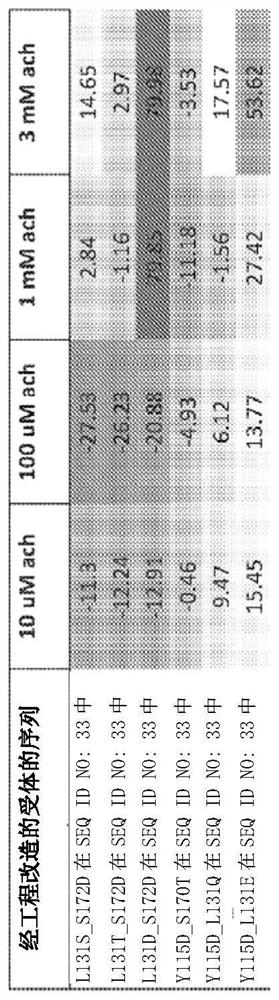
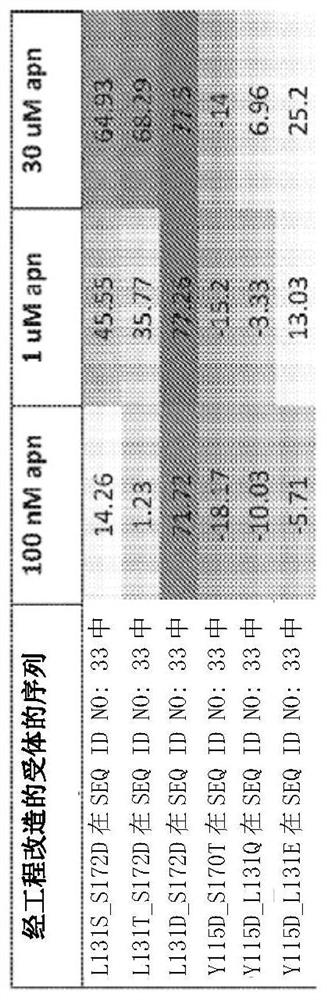
[0372] Example 1. Discovery of Mutated Engineered Receptors Containing a Ligand Binding Domain
[0373] To generate LGICs that conduct anion currents upon exposure to unnatural small-molecule agonists of human α7-nAChR, chimeric ligand-gated ion channel (LGIC) receptors were genetically engineered, containing receptors derived from human α7 nicotinic acetylcholine receptors. A ligand-binding domain (α7-nAChR) derived and a chloride-conducting ionopore domain derived from human GlyR1α. An engineered receptor with the amino acid sequence of SEQ ID NO: 33 was identified that was approximately as sensitive to acetylcholine, ABT-126, and TC-6987 as wild-type α7-nAChR, with TC-6987 showing similar Partial agonist activity on SEQ ID NO:33. Compared to wild type, SEQ ID NO:33 was about 2-fold less sensitive to nicotine and about 3-fold and 10-fold more sensitive to AZD-0328 and Facinicline / RG3487, respectively.
[0374] Amino acid substitutions were introduced into the ligand bindin...
Embodiment 2
[0376] Example 2: Characterization of engineered receptors using high-throughput fluorescence-based plate screening
[0377]To screen these mutant LGICs for those with novel responsive properties to ligands, an anion reporter assay was developed to assess channel function in a high-throughput format. In this assay, cells expressing a YFP reporter whose fluorescence is quenched in the presence of anions are transfected with DNA encoding the channel of interest. Upon exposure to the ligand, the activated channel will efflux anions, resulting in a dose-dependent quenching of YFP detectable on a plate reader.
[0378] Lenti-X 293T cells (LX293T, Clontech) were maintained in DMEM (Invitrogen) containing 10% FBS and 1% penicillin / streptomycin. For the plate reader assay, LX293T cells were infected with lentivirus to establish cells stably expressing the mutant YFP (H148Q / I152L) reporter, which showed enhanced sensitivity to anions. Two days before the assay, cells were aliquoted i...
Embodiment 3
[0382] Example 3: Characterization of engineered receptors using high-throughput electrophysiology
[0383] To confirm EC determined by plate reader 50 As well as a better understanding of the formation of maximal currents, a high-throughput electrophysiology system is performed on engineered receptors as described below. For HEK293T studies, the cDNA encoding the ion channel was cloned into pcDNA3.1 using standard recombinant techniques. HEK293T cells from Clontech (Lenti-X TM 293T cell line) were grown to 40-50% confluency in DMEM supplemented with 10% FBS and 1% penicillin / streptomycin and transfected with ion channel plasmids at a concentration of 18 μg per 15 cm dish using Fugene 6 and grown Another 24 hours. Cells were then assayed on an electrophysiology system (IonFluxHT and / or Mercury, Fluxion Biosciences), where dose-response relationships could be assessed by a microfluidic-based platform for establishing whole-cell configurations. with extracellular buffer (140...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com