Method for regulating methylation level of plant genome DNA (Deoxyribose Nucleic Acid) specific region
A plant genome and specific region technology, which is applied in the field of regulating the methylation level of specific regions of plant genome DNA to achieve the effect of ensuring food security
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
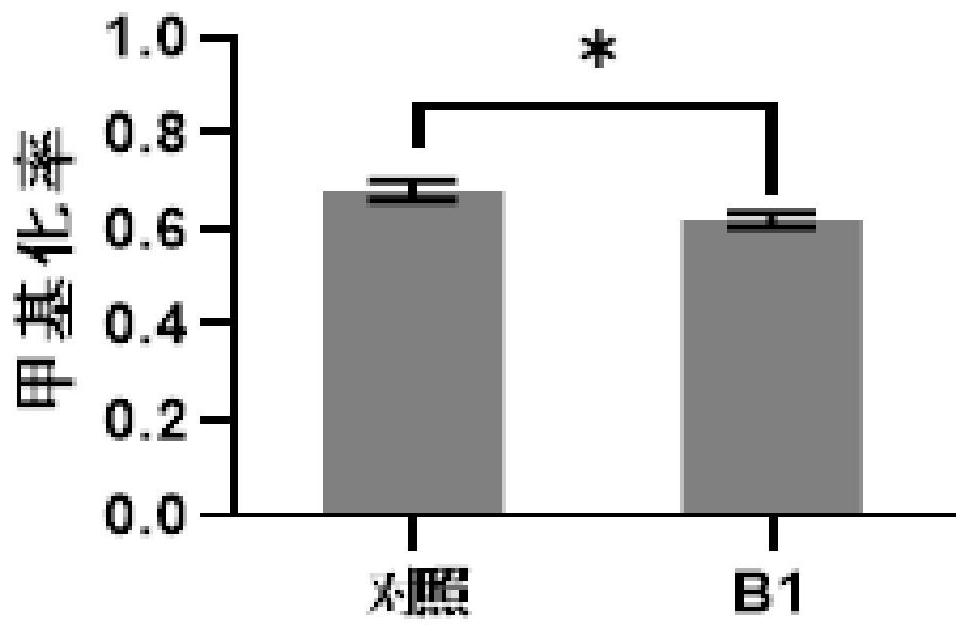
[0076] Example 1. Regulation of methylation levels in specific regions of rice genomic DNA
[0077] 1. Construct A1 and Construct A2
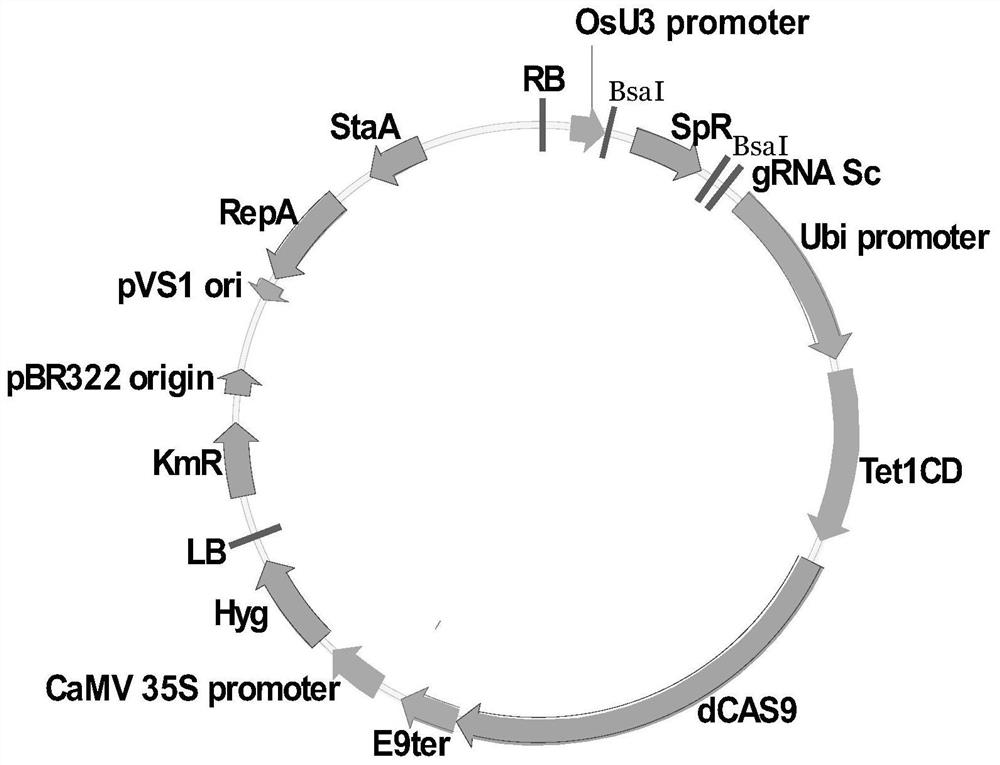
[0078] The plasmid map of construct A1 is as follows figure 1 As shown, construct A1 is an expression construct for reducing the methylation level of the target and nearby regions, which can express Tet1CD-dCas9 fusion protein, the nucleic acid sequence of Tet1CD-dCas9 is shown in SEQ ID No. As shown in SEQ ID No.12. Positions 1-741 of SEQ ID No. 12 are Tet1CD domains (encoding nucleic acids are positions 1-2223 of SEQ ID No. 11); positions 742-2243 are dCas9 domains (encoding nucleic acids are positions 1-2223 of SEQ ID No. 11) of bits 2224-6732). The original vector of this construct is the conventional CRISPR / Cas9 vector for rice transformation (pH-nCas9-PBE, described in "Zong, Yuan, et al." Precise base editing inrice, wheat and maize with a Cas9-cytidine deaminase fusion." Naturebiotechnology 35.5(2017):438." article), construct A1 wa...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap