Method for predicting microbial population sensing gene abundance
A quorum sensing and microorganism technology, applied in the field of microorganisms, to achieve the effect of enriching annotation results and improving annotation efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
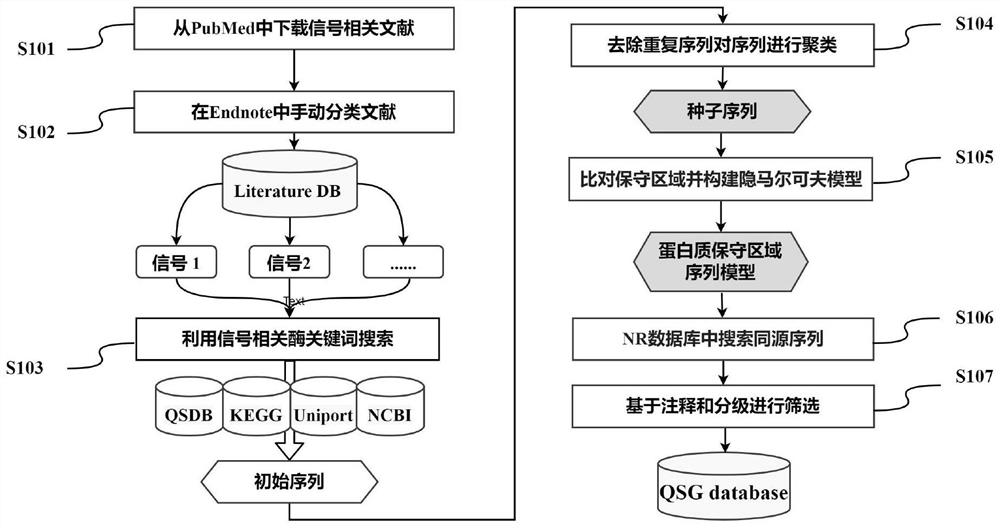
[0063] like figure 1 As shown, the present invention constructs a database process, and the specific steps are as follows:
[0064] S101. Search all literatures related to quorum sensing in the PubMed database of the National Center for Biotechnology Information (NCBI).
[0065] For the quorum sensing literature library involved in the present invention, all literatures related to the keyword "quorum" were retrieved from the PubMed database in NCBI, and as of October 30, 2021, a total of 12,596 literatures were collected.
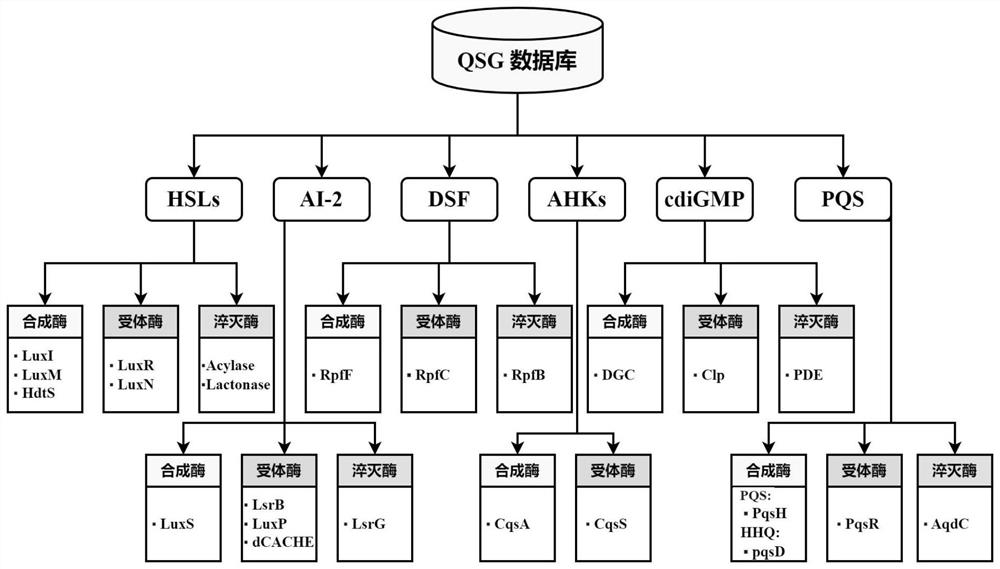
[0066] S102, using Endnote software to manually classify the literature, and finally form a quorum sensing literature library based on signals and corresponding genes, including 6 kinds of signal molecules and 24 kinds of related genes related to them.
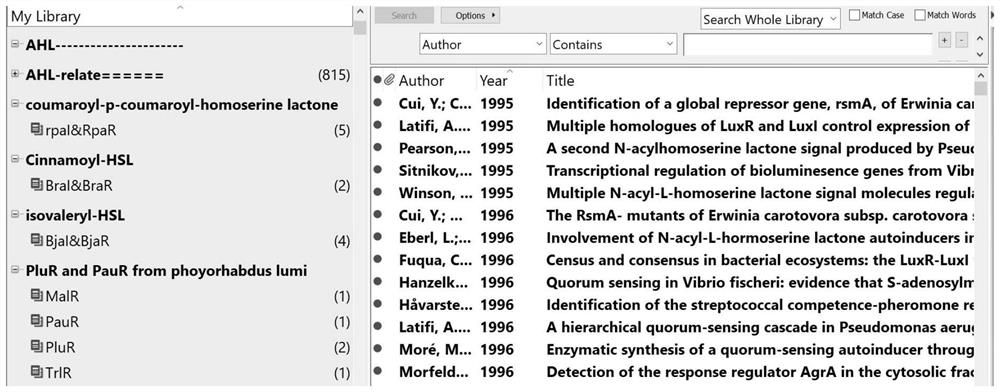
[0067] as attached figure 2 As shown in the screenshot of the document index database of the present invention, after removing 2167 irrelevant documents and classifying them according to signal molecules...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com