Homoeotic box gene 9Brn-1 from quail and its use
A homology, amino acid technology, applied in the field of genetic engineering, can solve problems such as less work in birds
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Embodiment 1: Molecular cloning and sequence determination of the cDNA of qBrn-1
[0026] Step 1: Preparation of Screening Library Probes
[0027] Chemically synthesize the following oligonucleotide sequences:
[0028] Primer 1: 5'CGACCTGGAGCAGTTCGCCAA 3'
[0029] Primer 2: 5'AACCAGACACGCACCACCT 3,
[0030] Quail embryos incubated for five days were taken, and cDNA was prepared by using mRNA purification Kit and cDNA synthesis Kit (Pharmacia Biotech) according to the method described in the manual. Using primer 1 and primer 2, use Taq DNA polymerase to carry out PCR reaction. After incubation at 97°C for 10 minutes, enter the following cycle: annealing at 55°C for 1 minute, extension at 72°C for 2 minutes, denaturation at 94°C for 1 minute, and cycle 35 times to obtain The DNA fragment of the screening library, which is cloned at the EcoRI site of the pBluescriptII SK-plasmid, has a total length of 402 bp, and its sequence is as follows:
[0031] 1 CGACCTGGAG CAGTTC...
Embodiment 2
[0045] Example 2: Determination of the restriction enzyme map of qBrn-1 cDNA and construction of various subclones
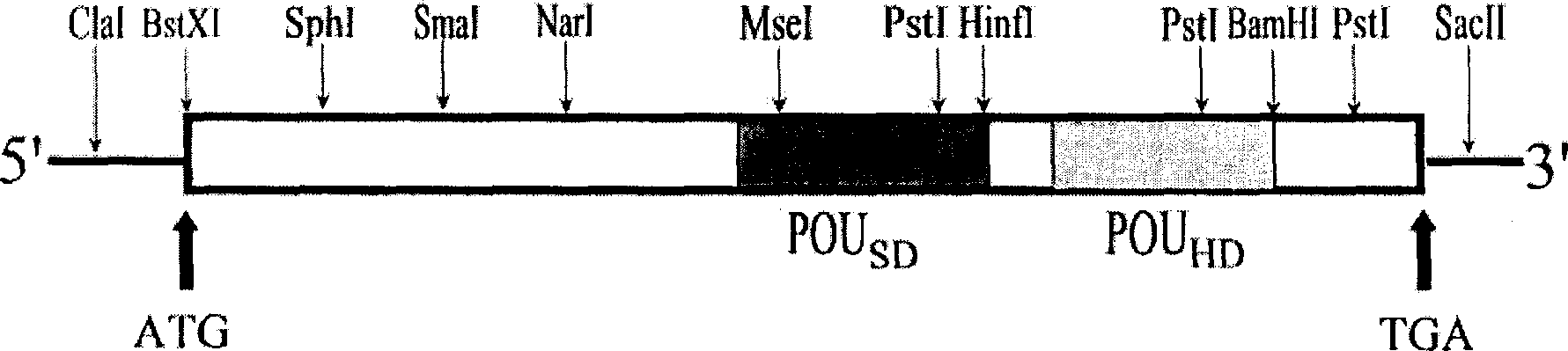
[0046] Step 1: Carry out single-digestion, double-digestion and multiple-digestion on the qBrn-1 clone of the present invention, and calculate the size of each digestion fragment by agarose gel electrophoresis, and analyze its multiple cloning site, and draw the restriction enzyme cut map, see figure 1 .
[0047] Step 2: qBrn-1 was double digested with Cla I and Sac I, and the digested fragment was recovered by agarose gel electrophoresis, and then pBluescript II KS was used as the carrier, and the same enzyme (ClaI and Sac I) was used It was double-digested, and finally subclone A was constructed by ligation reaction; in the same way, qBrn-1 was double-digested with BamHI and EcoR I to construct subclone B; qBrn-1 was double-digested with EcoRI and Cla I Digested with restriction enzymes to construct subclone C; single digested qBrn-1 with Pst I to construct ...
Embodiment 3
[0048] Embodiment 3: the preparation of antisense nucleic acid probe
[0049] The gene fragments in subclones B, C and D were selected as templates to prepare various antisense nucleic acid probes. The various subclones of the polynucleotide molecules of the present invention are shown in the accompanying drawings in Example 2, and the purpose of selecting three kinds of probes is to mutually verify the reliability of the results. The specific preparation method includes selecting an appropriate restriction enzyme to linearize the circular recombinant vector (i.e. subclone B, C and D), the restriction enzyme cutting site is located in the cDNA front of qBrn-1 Then use the polymerase (T3 polymerase or T7 polymerase) corresponding to the upstream of the 3' end of the positive strand of the cDNA to transcribe the linearized template in vitro, and the transcription product is the desired antisense nucleic acid probe.
[0050] In the present invention, in the in vitro transcriptio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com