Oligonucleotide micro-array chip for detecting small molecule RNA
A microarray chip and antisense oligonucleotide technology, applied in the biological field, can solve the problems such as the difference and interference of miRNA abundance that cannot be accurately reflected, and achieve the effects of large dynamic range, low cost and high sensitivity of detection.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
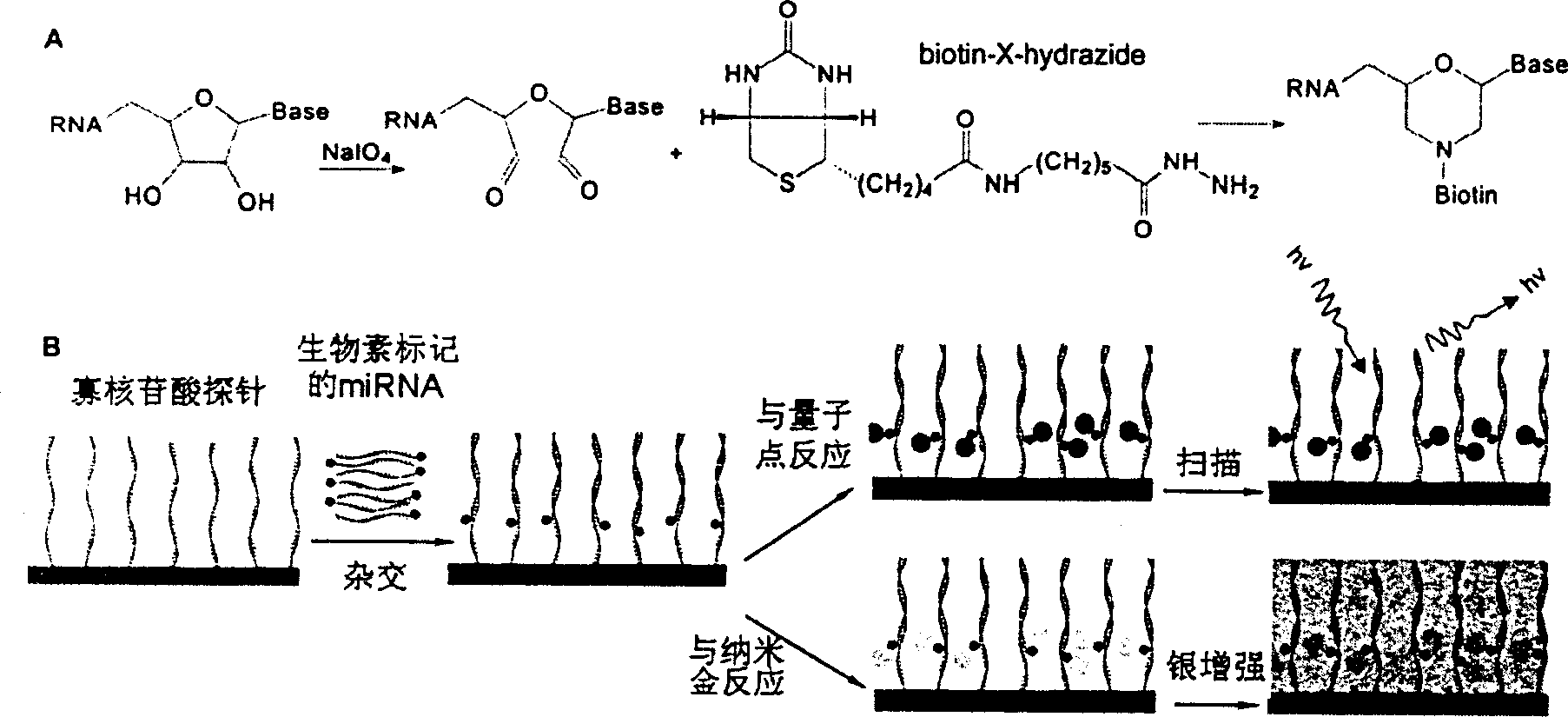
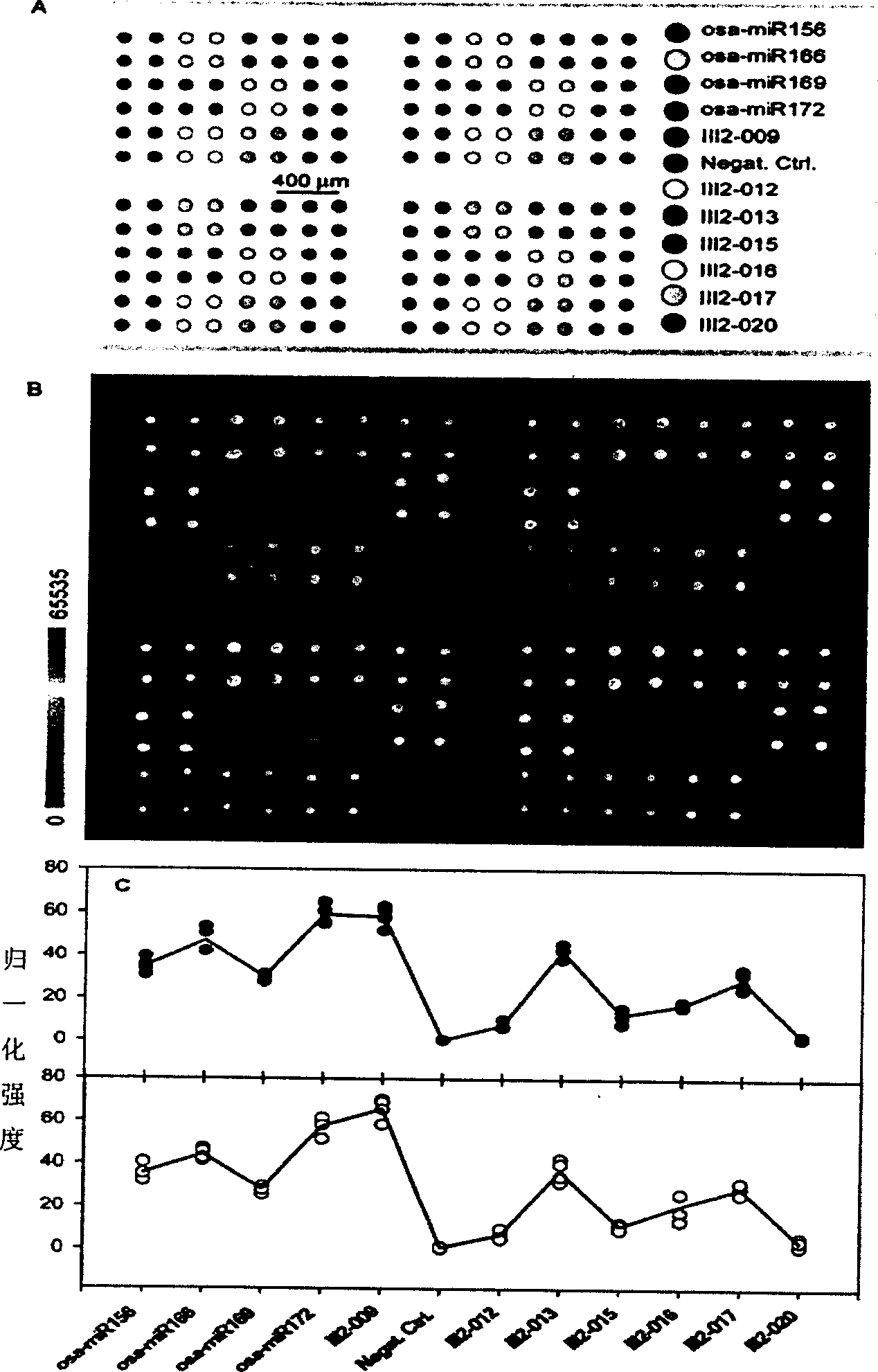
[0023] 11 antisense oligonucleotides (such as figure 2 A) and a negative control (Negat.Ctrl.) that is not complementary to known miRNAs were used to make miRNA detection microarray chips. The RNA fraction enriched with small molecular weight was first oxidized with sodium periodate, and then reacted with biotin-X-hydrazide (purchased from Sigma). After the reaction, the biotin-labeled RNA was precipitated with ethanol. The labeled RNA was hybridized with the detection chip in a hybridization solution (50% formamide prehybridization / hybridization solution at 25° C. for 18 hours), and then reacted with streptavidin-coupled quantum dots. After washing (in 1×SSC / 0.5% SDS at 37° C. for 10 minutes), scan with ScanArray 5000, and then perform normalization analysis with QuantArray. The result is as figure 2 Shown in B and C.
example 2
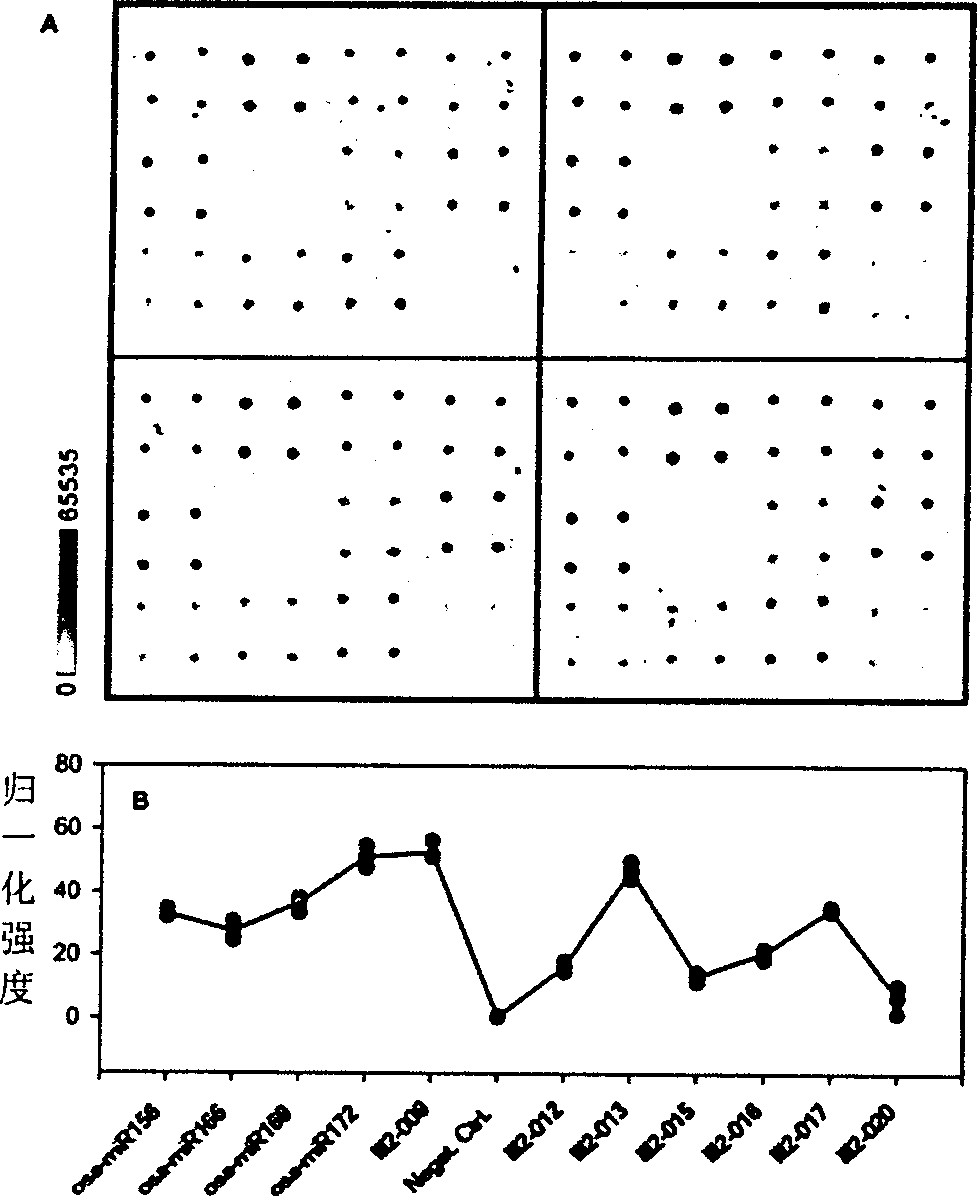
[0025]The labeled RNA obtained in step (1) was hybridized with the detection chip in the hybridization solution, and then reacted with colloidal gold (purchased from Sigma) coupled with streptavidin (colloidal gold was diluted 1:5, and 10 Microliter reacted with the chip, room temperature for 1 hour), followed by silver enhancement for 20 minutes. Take pictures with a common digital camera after washing, then analyze with QuantArray, the results are as follows: image 3 shown.
[0026] From figure 2 and image 3 It can be seen from the results that both methods give the same miRNA expression profile, and both have good repeatability. From Figure 4 It can be seen from the results that the detection limit of the miRNA microarray using the labeling method of the present invention can reach 39 pmol. Furthermore, the inventors believe that under optimal conditions the limit of detection can be lower.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com