Preparation method of T vector for T-A cloning
A system, T-A technology, applied in the direction of using vectors to introduce foreign genetic material, recombinant DNA technology, etc., can solve the problems of difficult quality control, expensive endonucleases, and high background of non-recombinant transformants
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
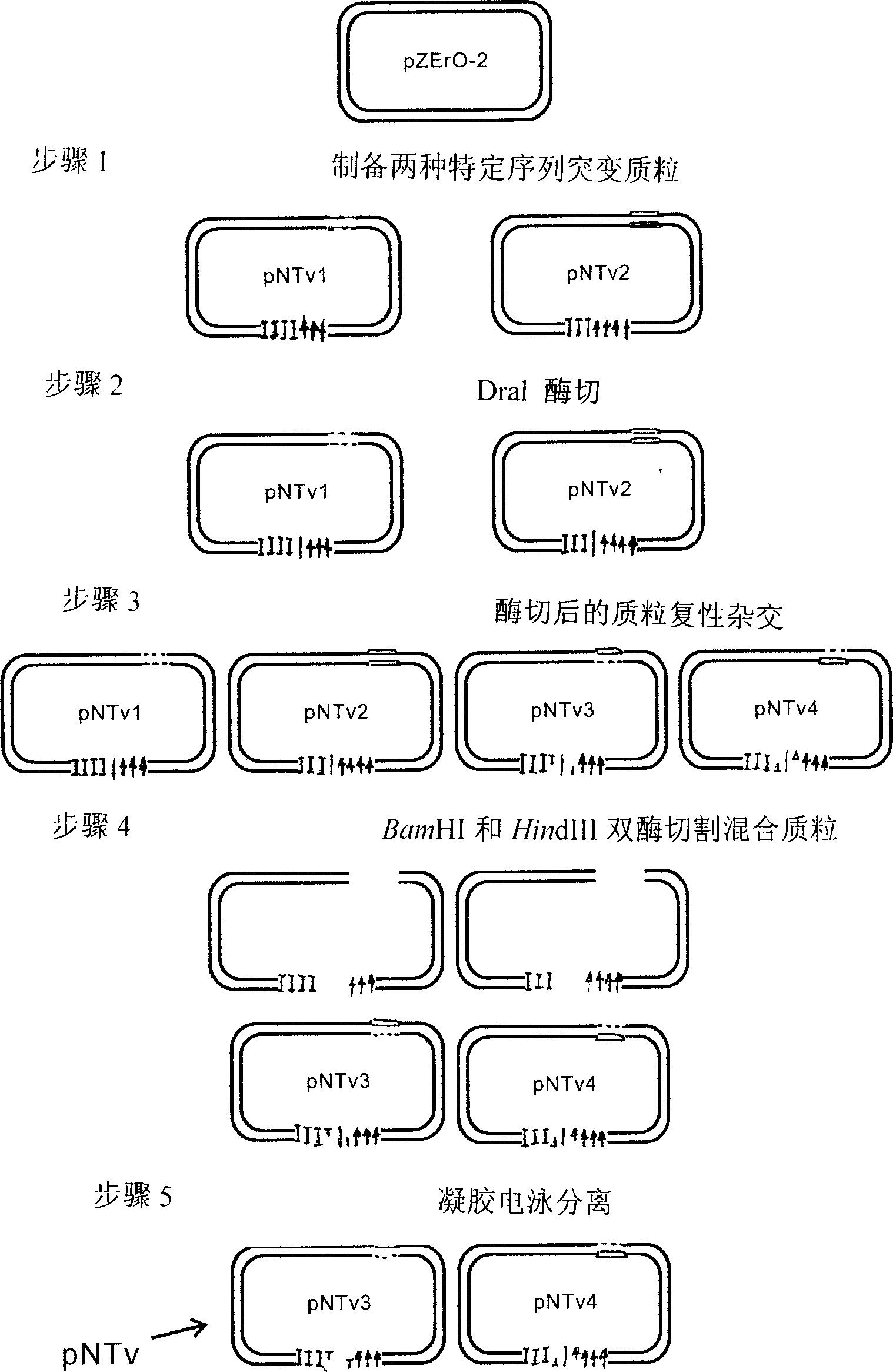
[0020] Embodiment 1: Introduce the T carrier preparation method that is used for T-A clone, as figure 1 :
[0021] Choose a commonly used plasmid with a high copy number, such as pBR322 series plasmids, pUC series plasmids, etc.
[0022] This example selects the pZErO-2 plasmid as a template.
[0023] 1. Using the pZErO-2 plasmid as a template, four pairs of primers were designed according to its DNA sequence, and two mutant plasmids with differences in specific sequences were constructed through site-directed mutagenesis. The two new plasmids were named pNTv1 and pNTv2 respectively. Among them, a new restriction site BamHI and a TTTTAAA sequence box that can be recognized by DraI endonuclease are introduced into pNTv1, as shown in Figure 1: the dashed line represents the restriction site BamHI, and the I and arrow symbols in the figure represent the TTTTAAA sequence box , can be cut by DraI endonuclease; pNTv2 introduces a new restriction site HindIII and a TTTAAAA sequence b...
Embodiment 2
[0029] Embodiment 2: Performance detection of T carrier pNTv
[0030] 1. The mixture of recovered pNTv3 and pNTv4 was mixed with DNA with 3′dA in three different molar ratios of 1:3, 1:5, and 1:7, and T4 DNA ligase was added for ligation at 16°C.
[0031] 2. The TOP 10 competent cells were electrotransformed with the ligated products.
[0032] 3. Spread the transformed product on LB plates and incubate at 37°C.
[0033] 4. PCR screening of colonies and extraction of plasmids for identification by enzyme digestion, the results showed that the recombination efficiency was above 90%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com