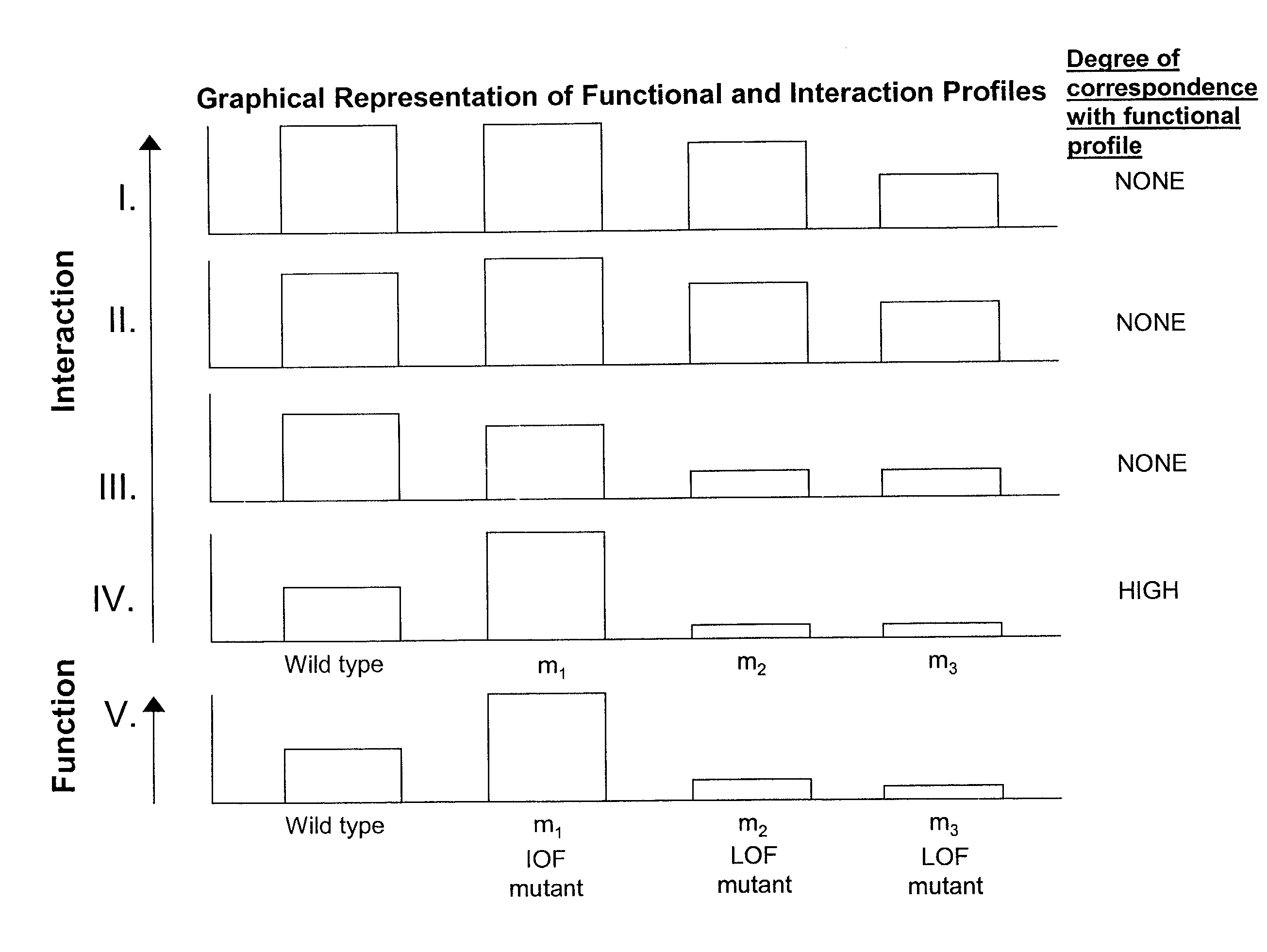
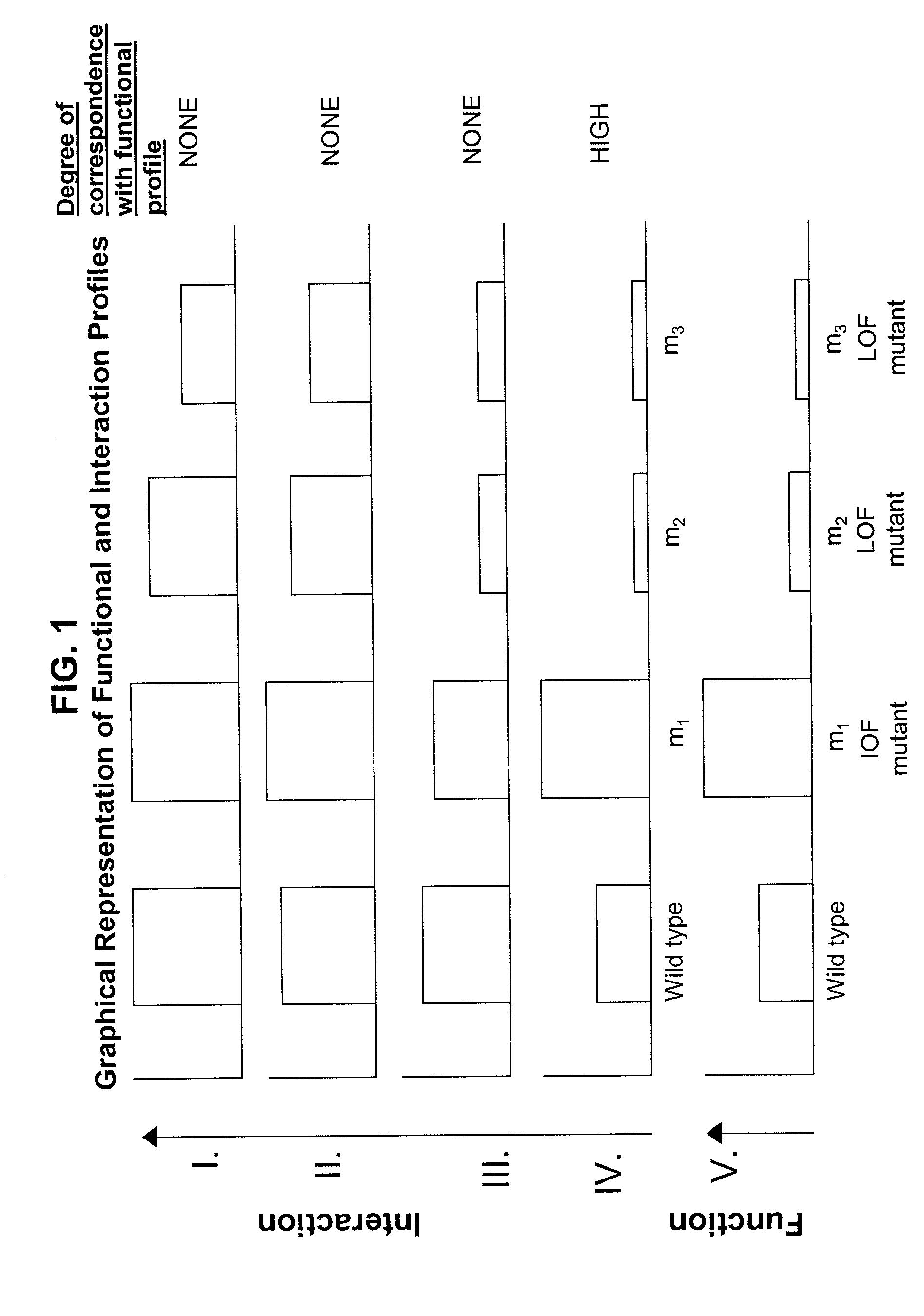
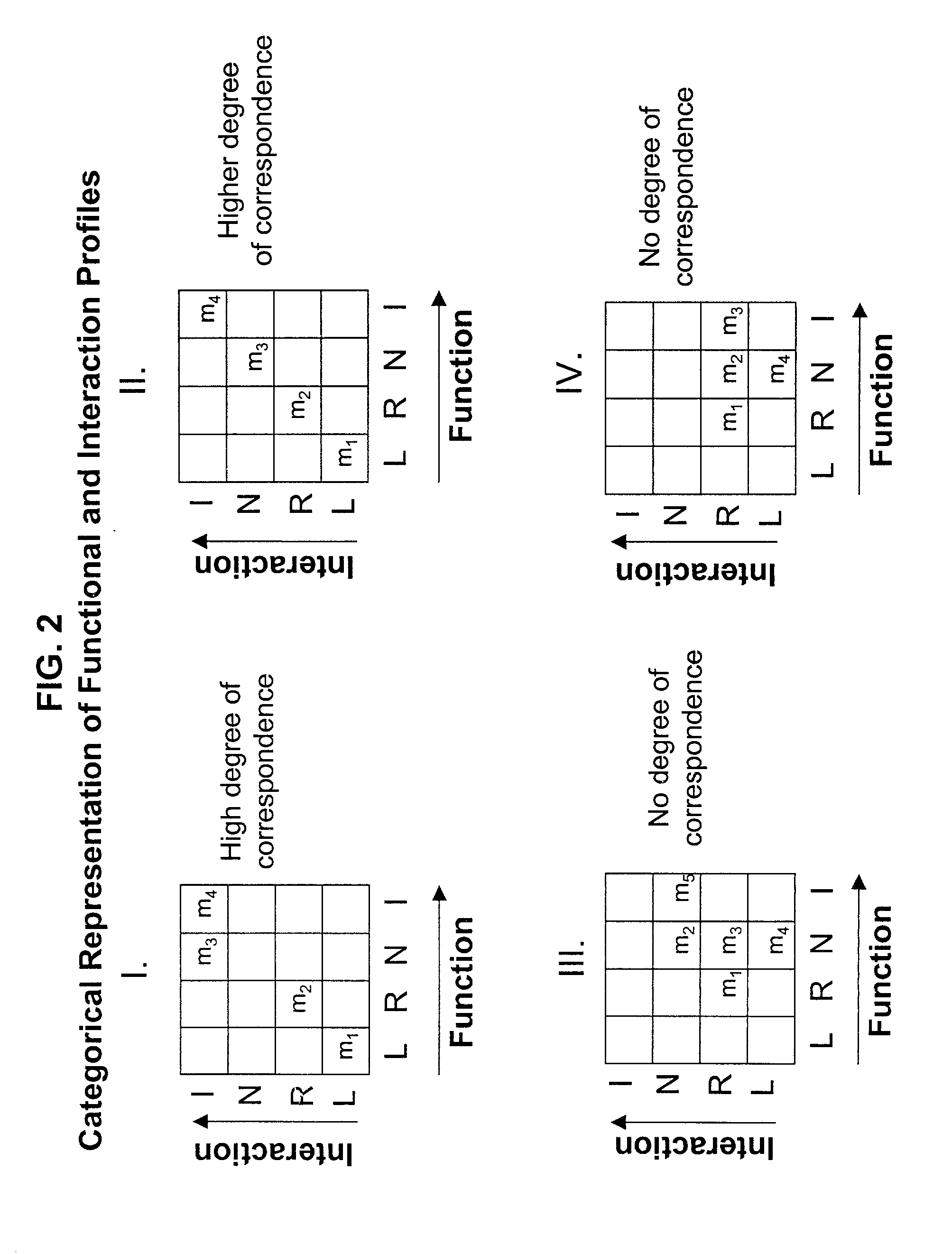
Methods of identifying target polypeptides
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0033] 5.1 Abbreviations
[0034] The abbreviations used for the genetically encoded amino acids are conventional and are as follows:
1 Amino Acid Three-Letter Abbreviation One-Letter Abbreviation Alanine Ala A Arginine Arg R Asparagine Asn N Aspartate Asp D Cysteine Cys C Glutamate Glu B Glutamine Gln Q Glycine Gly G Histidine His H Isoleucine Ile I Leucine Leu L Lysine Lys K Methionine Met M Phenylalanine Phe F Proline Pro P Serine Ser S Threonine Thr T Tryptophan Trp W Tyrosine Tyr Y Valine Val V
[0035] When the three-letter abbreviations are used, unless specifically preceded by an "L" or a "D," the amino acid may be in either the L- or D-configuration about .alpha.-carbon (C.sub..alpha.). For example, whereas "Ala" designates alanine without specifying the configuration about the .alpha.-carbon, "D-Ala" and "L-Ala" designate D-alanine and L-alanine, respectively. When the one-letter abbreviations are used, upper case letters designate amino acids in the L-configuration about the .al...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com