Encoded self-assembling chemical libraries (ESACHEL)
a chemical library and encoded technology, applied in the field of encoded self-assembling chemical libraries, can solve the problems of limited to this special type of chemical moiety, and the disadvantage of individual synthesis for each individual of a chemical library, and achieve the effect of more thermodynamic stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0094] As mentioned in previous sections, one strength of ESACHEL technology is its compatibility with a variety of different chemical moieties, including peptides and globular proteins (e.g., antibody domains).
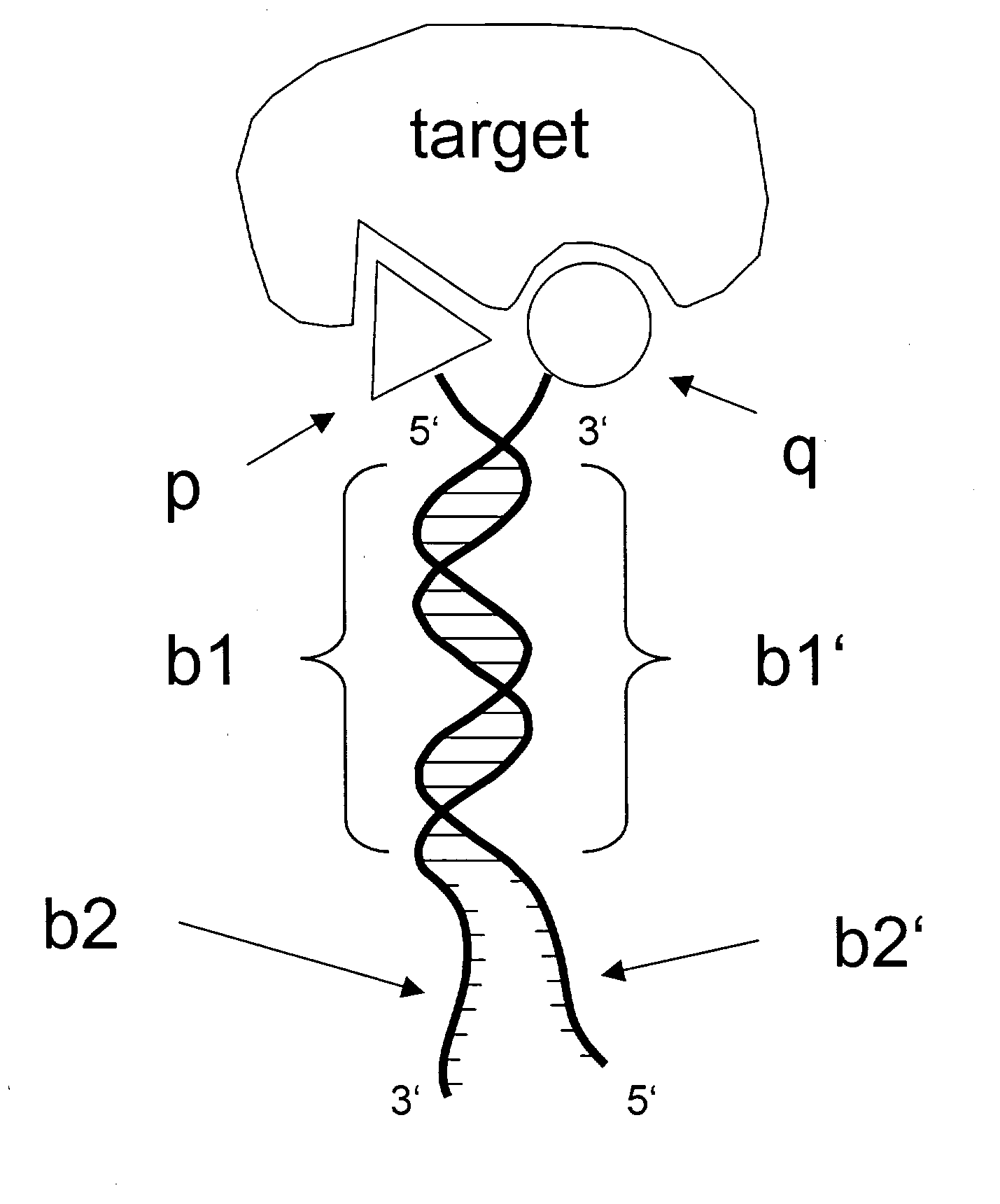
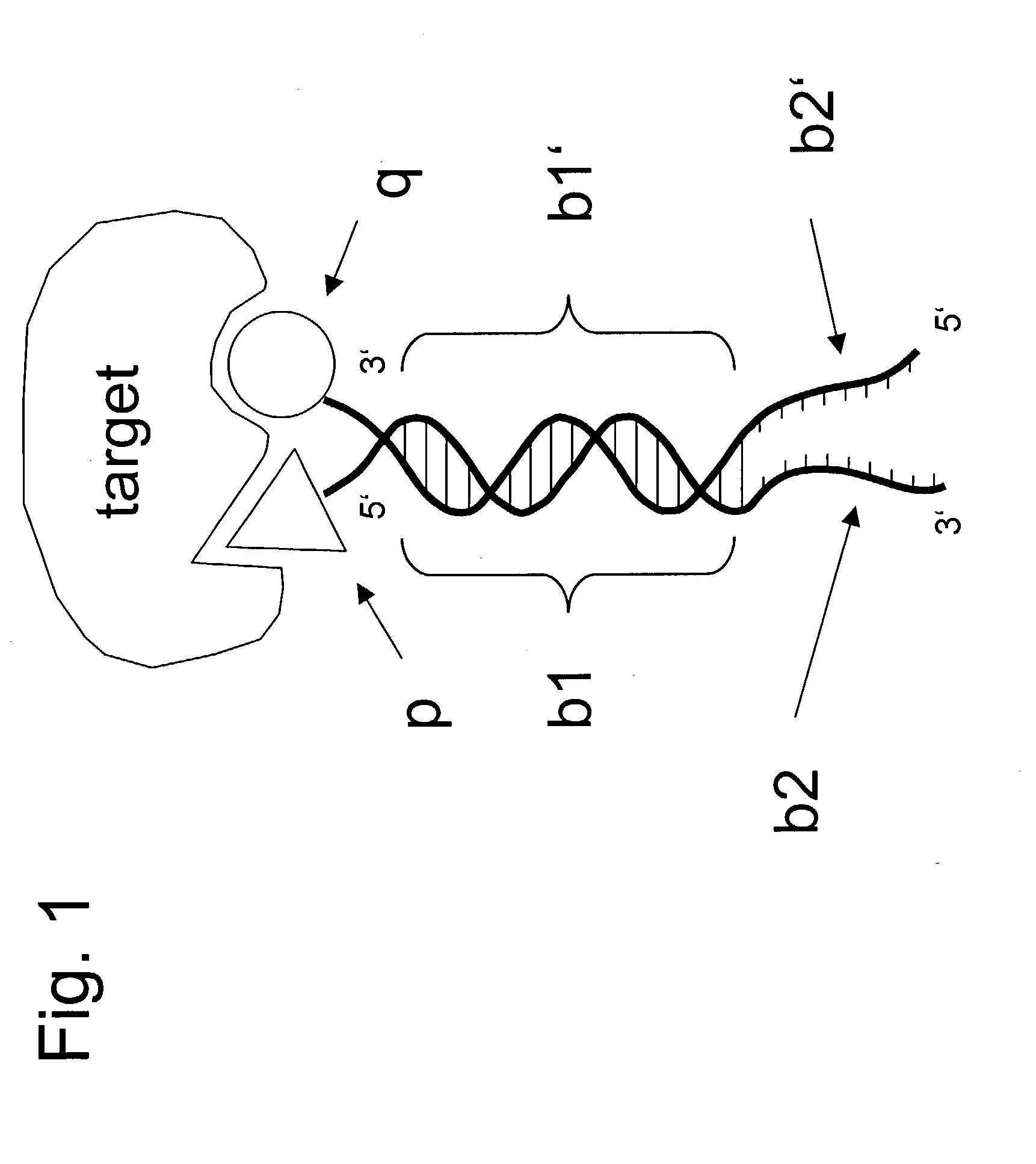
[0095] In this example, we show how a simple embodiment of ESACHEL (FIG. 1), featuring cysteine-tagged antibody variable domains covalently linked to DNA oligonucleotides capable of partial heteroduplex formation, leads to the identification of a pair of variable heavy domain (VH) and variable light domain (VL), which yield a specific antigen binding after heterodimerization.
[0096] The genes of the VH and VL domains of the L19 antibody (specific for the ED-B domain of fibronectin [Pini A, Viti F, Santucci A, Carnemolla B, Zardi L, Neri P, Neri D. Design and use of a phage display library. Human antibodies with subnanomolar affinity against a marker of angiogenesis eluted from a two-dimensional gel. J Biol Chem. (1998) 273:21769-21776]), of the HyHEL-10 antibody (specific for ...
example 2
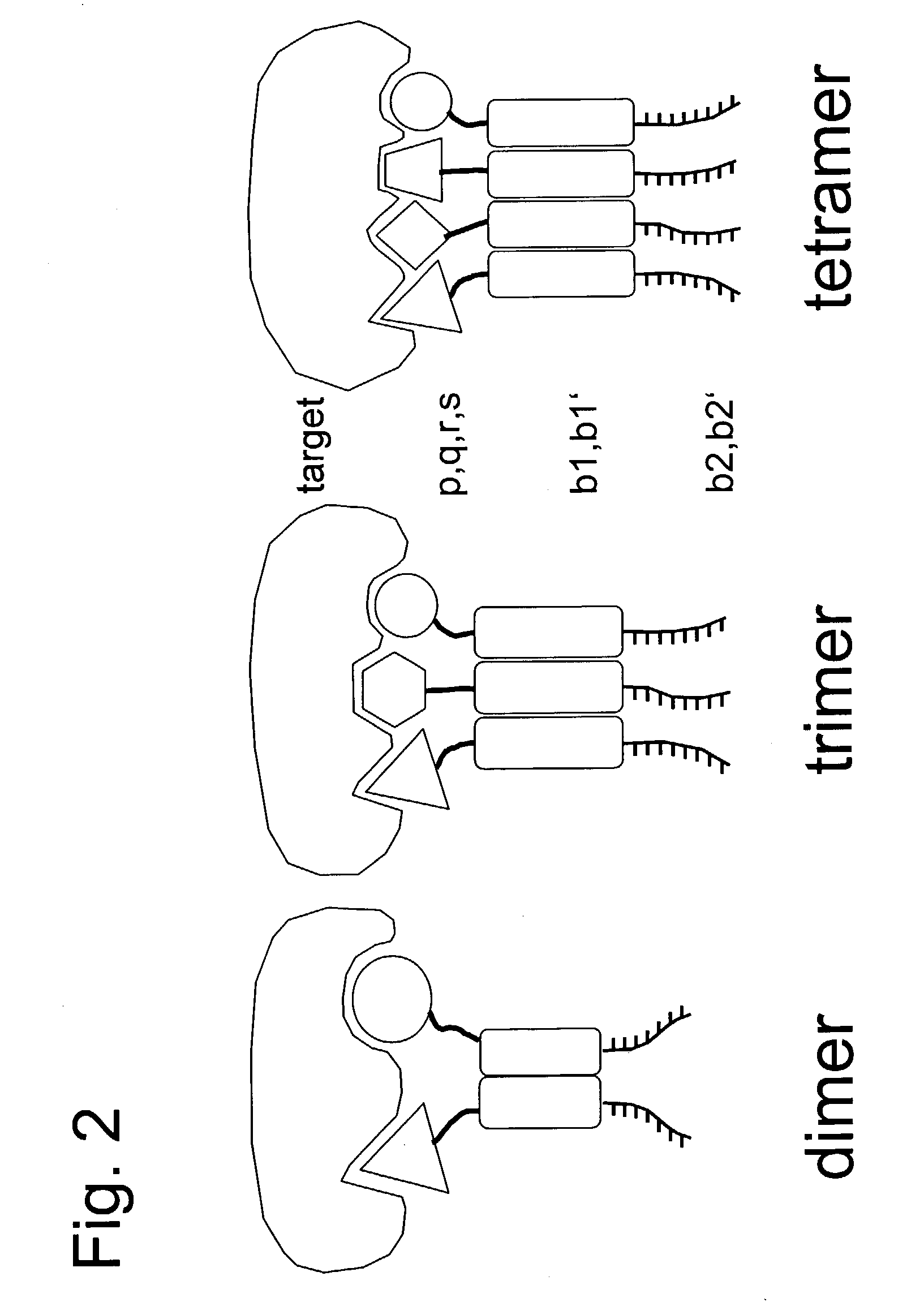
[0106] In this example, we describe how the ESACHEL embodiment of FIG. 1 can be performed in a practical implementation. The experimental strategy outlined here is also applicable to the embodiments described in FIG. 4, in which DNA triplexes or DNA quadruplexes are used to display chemical entities at the extremity of self-assembling oligonucleotides.
[0107] Two sub-libraries are constructed as follows: A sub-library "A" is created, by coupling n compounds to the 3' extremity of n different DNA oligonucleotides. Among the many different possible implementations, a convenient one is represented by the coupling of iodoacetamido- or maleimido-derivatives of n chemical entities to individual DNA oligonucleotides, which carry a thiol group at the 3' end. The coupling can easily be performed at room temperature in PBS (50 mM phosphate buffer+100 mM NaCl, pH=7.4), by simple mixing of the thiol-bearing oligonucleotide (typical concentration range: 10-100 .mu.M) with a molar excess of iodoac...
example 3
[0111] This example illustrates one of the many possible decoding methodologies, for ESACHEL embodiments as described in FIG. 1 and in Example 2.
[0112] The decoding strategy, schematically depicted in FIG. 5, is based on the principle that, after biopanning of desired ESACHEL binding specificities, PCR fragments are generated, each of which carries the code of pairs of sub-library members, whose combination was rescued in the biopanning experiment, therefore allowing the identification of the corresponding heterodimerized chemical entities.
[0113] Chemical entities of sub-libraries A and B (see also FIG. 1 and Example 2) are coupled, individually, to members of two pools of DNA oligonucleotides with the following properties:
[0114] One pool of oligonucleotides carries the chemical entities at the 3'-end (pool A), whereas the other pool carries the chemical entity at the 5'-end (pool B).
[0115] A sufficient number of bases at the 5' extremity of oligonucleotides of pool B allow the spec...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com