Mismatch repair detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
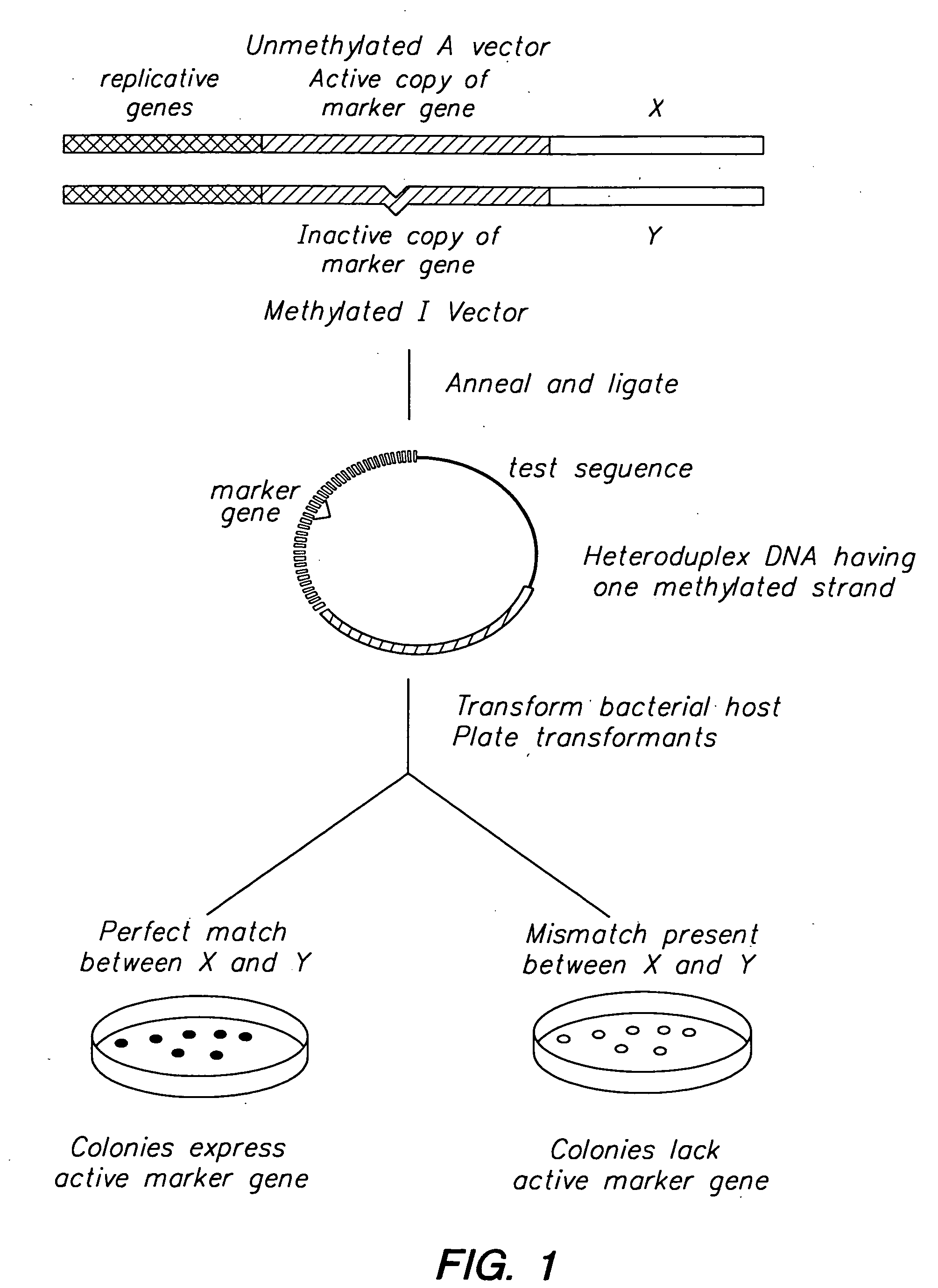
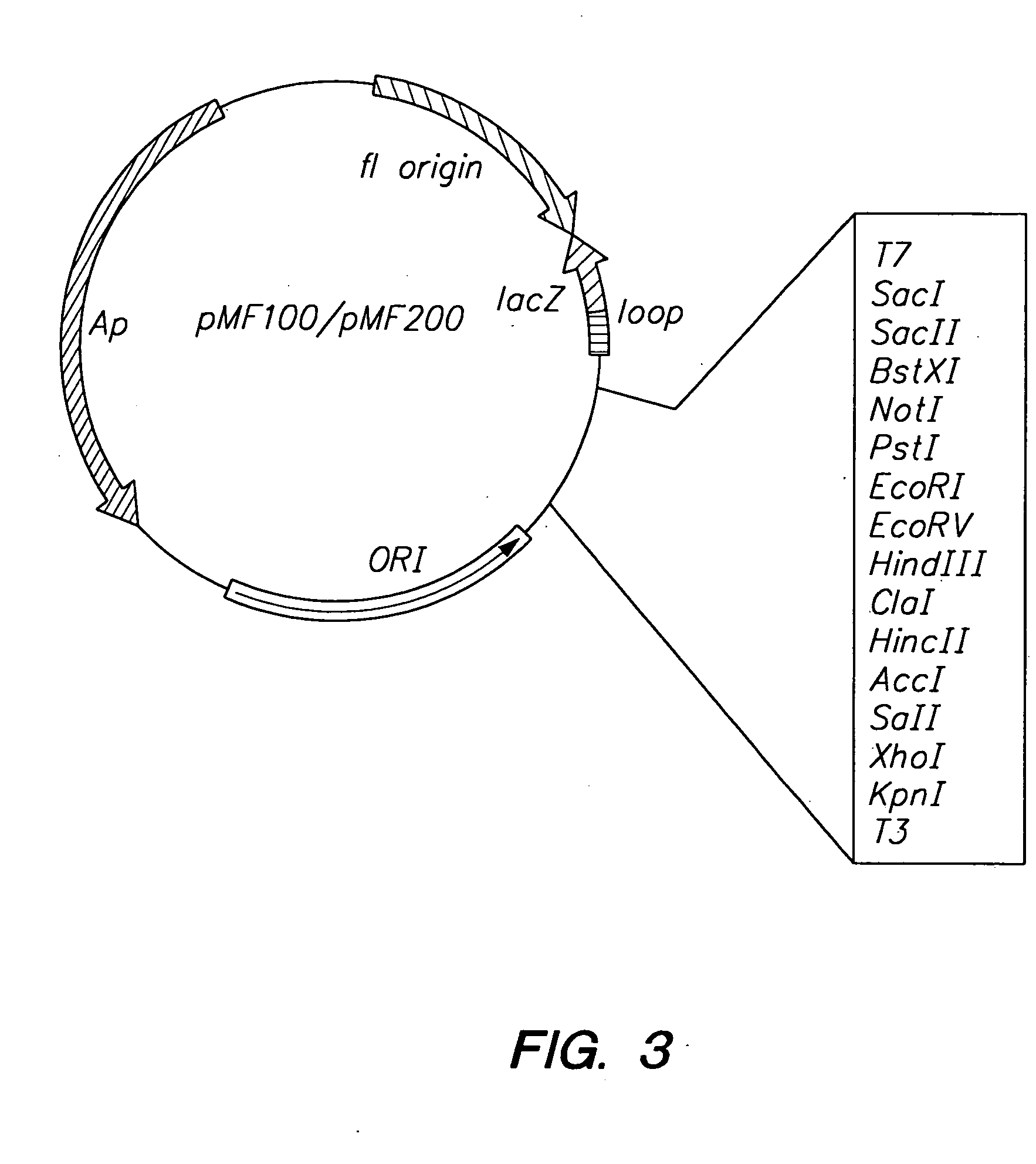
[0093] Two pUC-derived plasmids, the A plasmid (pMF200) and the I plasmid (pMF100), are employed in the MRD procedure. A map of the plasmids is shown in FIG. 3. These plasmids are identical except for a five bp insertion into the Lac Zα gene of pMF100. This insertion results in white colonies when bacteria transformed with the I plasmid are grown on LB plates supplemented with indolyl-β-D-galactoside (Xgal) and isopropyl-β-D-thiogalactoside (IPTG). In contrast, bacteria transformed with the A plasmid result in blue colonies when grown under these conditions.
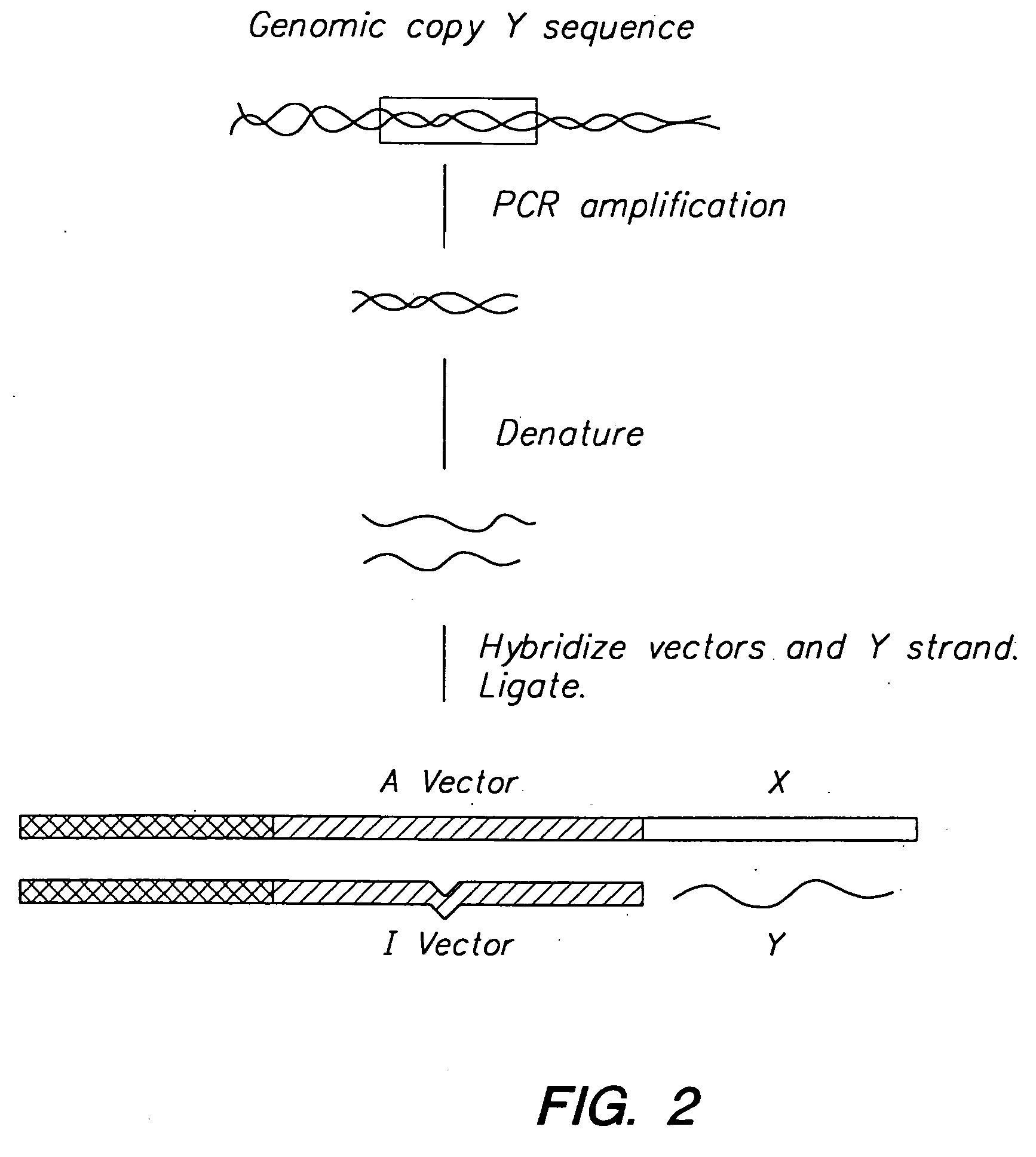
[0094] The initial step of the MRD procedure consists of cloning one of two DNA fragments to be screened for differences into the A plasmid and cloning of the second DNA fragment into the I plasmid. The A plasmid construct is then transformed into a dam− bacterial strain, resulting in a completely unmethylated plasmid while the I plasmid construct is transformed into a dam+ bacterial strain, resulting in a fully methylated plasm...
example 2
[0098] As an initial test of the sensitivity and specificity of the MRD system, a single nucleotide mismatch was detected in a 550 base pair DNA fragment derived from the promoter of the mouse beta globin gene (Myers et al. (1985) Science 229:242). MRD was used to compare this DNA fragment, which contains a T at position −49 (relative to the functional transcription start site of the gene) with a second DNA fragment identical in sequence except for at C position −49. The mismatch was located about 700 base pairs from the five nucleotide Lac Zα loop in the vector. Comparison of the two DNA molecules by using MRD resulted in 90% white colonies. In contrast, comparison of the same two DNA molecules with no mismatch (−49T / −49T), resulted in only 7% white colonies. The data is shown in Table 1.
TABLE 1Detection of Known Point Mutations using MRDSequenceDistance from% White (Inactive)Variation*Fragment Size{circumflex over ( )}Loop{circumflex over ( )}Colonies@None10.55N / A7G_C10.550.789A...
example 3
[0103] MRD was used to detect unknown mutations in genomic DNA fragments generated by the polymerase chain reaction (PCR). PCR is a practical method for obtaining a particular genomic DNA fragment of interest from many different individuals. Recent advances in PCR technology makes it possible to isolate DNA products greater than 10 kb in length (Barnes (1994) P.N.A.S. 91:2216; Cheng et al. (1994) P.N.A.S. 91:5695). However, the introduction of errors during the PCR reaction severely limits the use of individual cloned PCR products. In an effort to overcome this limitation, an MRD protocol was developed to enrich for molecules that are free of PCR-induced errors. Following this “cleaning” protocol, the cloned PCR products can be compared for DNA sequence differences by using the MRD procedure described above.
[0104] The basic principle underlying the MRD cleaning protocol is the fact that any single PCR-induced mutation will make up a very small fraction of all the molecules generate...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Electrical resistance | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com