Methods for isolating and characterizing endogenous mRNA-protein (mRNP) complexes
a technology of mrna-protein complexes and complexes, applied in the field of posttranscriptional regulation and methods of profiling gene expression, can solve the problems of inability to report on in vitro applications, and inability to independently assess each process en mass
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
RNase Protection in a Multiprobe System: Materials and Methods
[0078] It has previously been reported that HuB (Hel-N1) immunoprecipitation, using a g10 epitope tag, resulted in the co-immunoprecipitation of a mRNA, which once amplified by RT-PCR and sequenced, was found to encode NF-M protein (Antic, 1999, supra). In this example, the same approach is expanded to using a multiprobe RNase protection assay to rapidly optimize the immunoprecipitation of several endogenous mRNA-protein (mRNP) complexes containing different mRNA-binding proteins. In the multi-probe system, many mRNAs, from mRNP pellets, can be assayed in a single lane of polyacrylamide gel.
[0079] Cell Culture and Transformation. Murine P19 embryonal carcinoma cells were obtained from the ATCC and maintained in monolayer culture using alpha.-MEM without phenol red (Gibco-BRL 41061-029) supplemented with 7.5% Bovine Calf Serum, 2.5% Fetal Bovine Serum (Hyclone) and 100U Penicillin / Streptomycin. Cells were grown in tissue...
example 2
RNase Protection in a Multiprobe System: Experimental Results
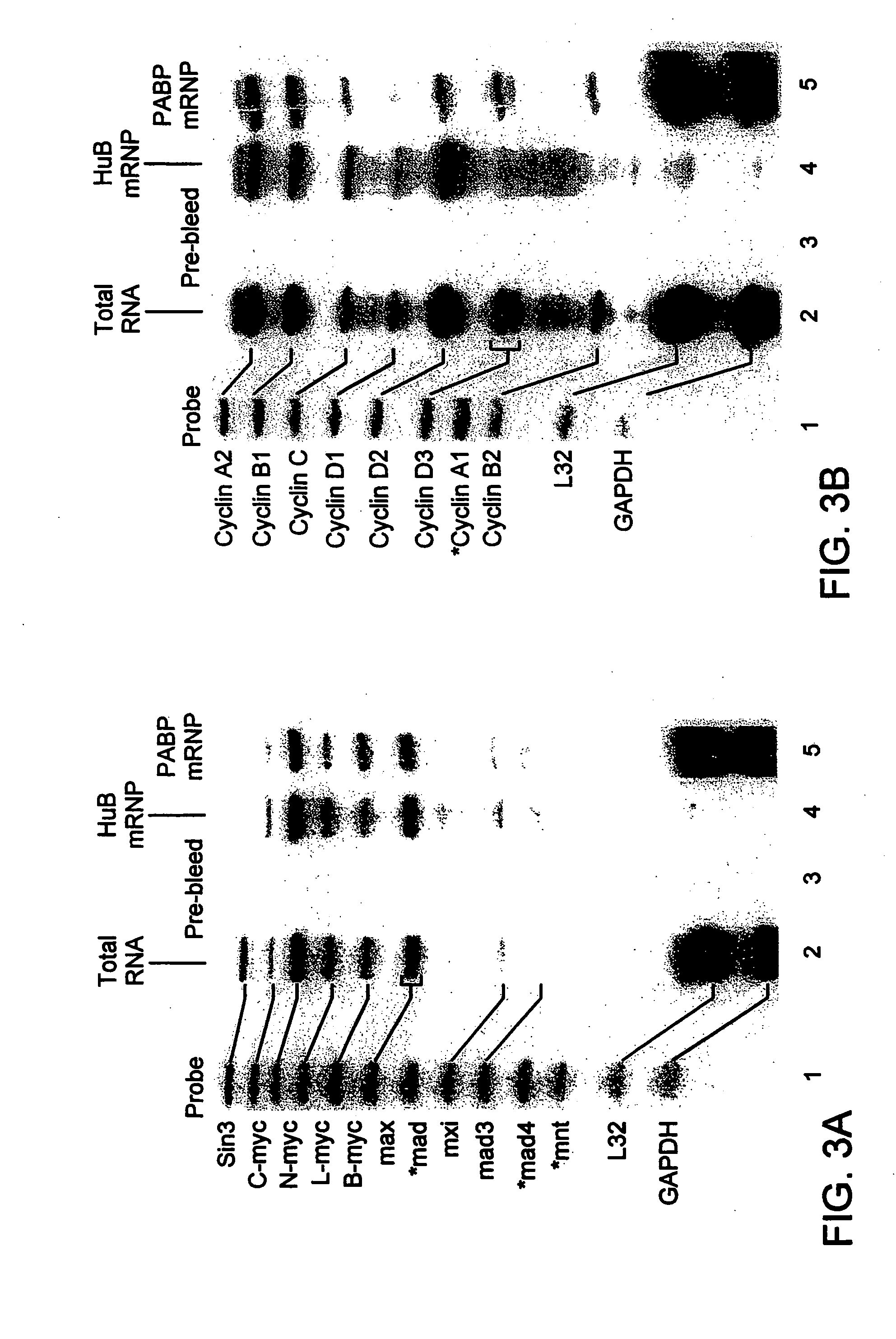
[0085]FIG. 3 shows an immunoprecipitation of HuB and Poly-A binding protein (PABP)-mRNP complexes from extracts of murine P19 cells stably transfected with g10-HuB cDNA. No mRNAs were detected in pellets immunoprecipitated with polyclonal pre-bleed rabbit sera (FIGS. 3A and 3B, lane 3), or with many other rabbit, mouse, and normal human sera tested with this assay (data not shown). The profiles of mRNAs associated with HuB mRNP complexes included n-myc, 1-myc, b-myc, max and cylins A2, B1, C, D1, and D2, but not sin3, cyclin D3, cyclin B2, L32 or GAPDH mRNAs (FIGS. 3A and 3B, lane 4). In contrast, the profiles of mRNAs extracted from PABP-mRNP complexes resembled the profiles of total RNA, but showed enriched levels of L32 and GAPDH and decreased levels of sin3 mRNA (FIGS. 3A and 3B, lane 5). It was concluded that antibodies reactive with these cellular RNA-binding proteins could be used to immunoprecipitate mRNP complexe...
example 3
Identification of mRNA Subsets Associated with RNA Binding Proteins En Masse Using cDNA Arrays: Materials and Methods
[0086] To further expand the ability to identify the mRNAs associated in endogenous mRNP complexes, this example describes the use of a cDNA array filter as a highly specific and sensitive method to detect a mRNA subset without amplification or iterative selection (FIG. 4).
[0087] Antibodies. Monoclonal anti-gene 10 (g10) antibodies were produced as previously described (see D. Tsai et al., Proc. Natl. Acad. Sci. USA, 89, 8864-8868 (1992); Gao et al. (1994) Proc. Natl. Acad. Sci. USA 91, 11207-11211; Antic et al. (1999) Genes Dev. 13, 449-461). Polyclonal sera reactive with Hu proteins were produced as previously described (Levine et al. (1993) Mol. Cell. Biol. 13, 3494-3504; Atasoy et al. (1998) J. Cell Sci. 111, 3145-3156). Antibody against 5′ cap binding protein (elF-4E) was obtained from Transduction Laboratories (San Diego, Calif.). Antibodies reactive with Poly...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com