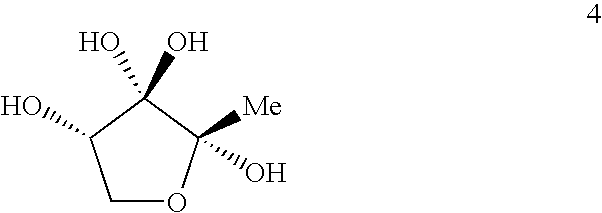AI-2 compounds and analogs based on Salmonella typhimurium LsrB structure
a technology of ai-2 and ai-2 compound, which is applied in the field of crystals comprising apolsrb and hololsrb, can solve the problems of inaccurate use of s-thmf-borate to refer exclusively to ai-2 and difficulty in identifying the active form of ai-2 signaling in v. harveyi, etc., and achieve the effect of inhibiting ai-2 activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Experimental Procedures
A. LsrB Production
[0091]S. typhimurium LsrB without its amino-terminal signal peptide (residues 1-26) was cloned into plasmid pGEX4T1 for expression as a glutathione-S-transferase (GST) fusion protein in E. coli strain BL21. Protein expression was induced by the addition of 0.1 mM isopropyl β-D-thiogalactopyranoside for 6 hr prior to harvesting the bacteria. The GST-LsrB fusion protein was purified by glutathione agarose affinity chromatography. The GST tag was removed by thrombin digestion, leaving two additional residues at the N terminus (GlySer) of LsrB. The protein was further purified by hydrophobic affinity chromatography (Phenyl Superose; Pharmacia) and size-exclusion chromatography (Superdex 200; Pharmacia). LsrB (>95% pure) was concentrated for crystallization experiments to 8 mg / ml in 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 1 mM dithiothreitol. Selenomethionyl protein was overexpressed in E. coli B834. Cells were grown in M9 medium as described...
example 2
Structure of LsrB
[0103] The structure of S. typhimurium LsrB was determined to 2.1 Å resolution using multiwavelength anomalous diffraction (MAD) phasing and subsequently refined to 1.9 Å resolution (Table 1). Despite low sequence identity (11%), LsrB exhibits the same fold as the V. harveyi AI-2 signaling receptor LuxP (Chen et al., 2002), with a three-stranded hinge connecting two similar α / β domains (FIG. 2A). LsrB also has strong structural homology with several other sugar binding proteins including E. coli ribose binding protein (RBP) and S. typhimurium galactose binding protein, as well as repressors such as E. coli purine nucleotide synthesis repressor and trehalose repressor (Hars et al., 1998; Mowbray et al., 1992; Mowbray et al., 1983; Schumacher et al., 1994). In periplasmic binding proteins, including LuxP, the ligand binding site is near the hinge between the two domains. In the crystal structure of LsrB, the domains are in an open conformation similar to, though less...
example 3
Structure of the LsrB:Ligand Complex
[0104] To identify the LsrB ligand, LsrB was crystallized in the presence of DPD and the other products generated by incubating SAH with recombinant Pfs and LuxS enzymes as previously described (Schauder et al., 2001) (FIG. 1A). The structure was determined by molecular replacement and refined to 1.3 Å resolution (Table 1). In this structure, the domains of LsrB have closed around the binding site, rotating shut about the hinge region by 21° relative to one another (FIG. 2B) (Hayward et al., 2002). Nonprotein electron density is prominent between the two domains (FIG. 2C) in a location analogous to the ligand binding sites of LuxP and other periplasmic binding proteins. As detailed below, this electron density is consistent with R-THMF, a DPD derivative not previously known to be biologically active (FIGS. 2D and 2E).
[0105] The LsrB ligand R-THMF differs from the V. harveyi LuxP ligand S-THMF-borate in two respects (FIG. 3A). First, no borate is...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com