Chimeric retroviral Gag genes and screening assays
a technology of chimeric retroviral and gag genes, applied in the field screening assays, can solve the problems of limiting the sequence of peptides to those containing the 20 genetically encoded amino acids, consuming time-consuming iterative procedures for searching for active sequences, and reducing the number of chimeric retroviral gag genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
M-PMV / HIV Chimeric Molecules
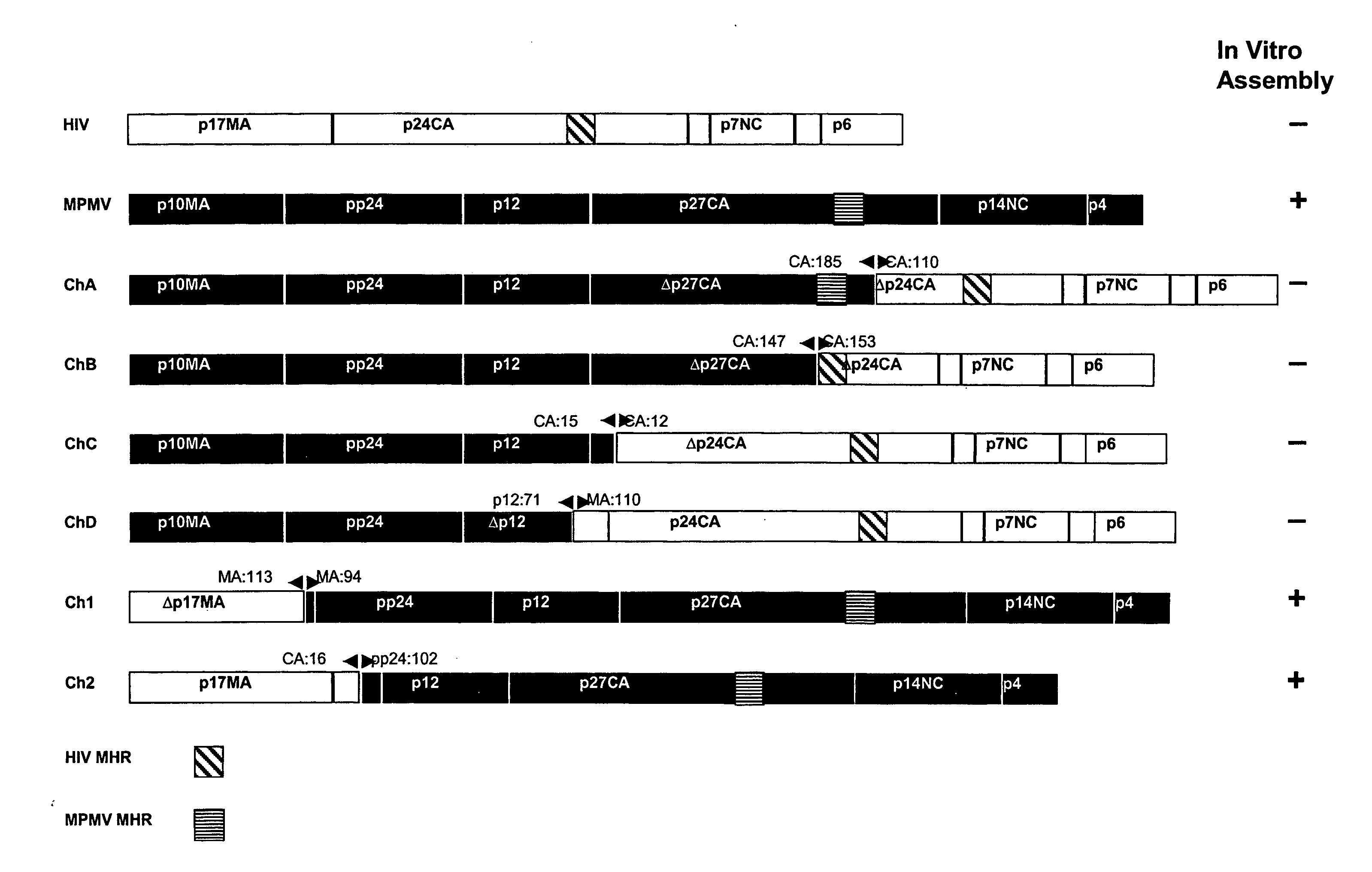
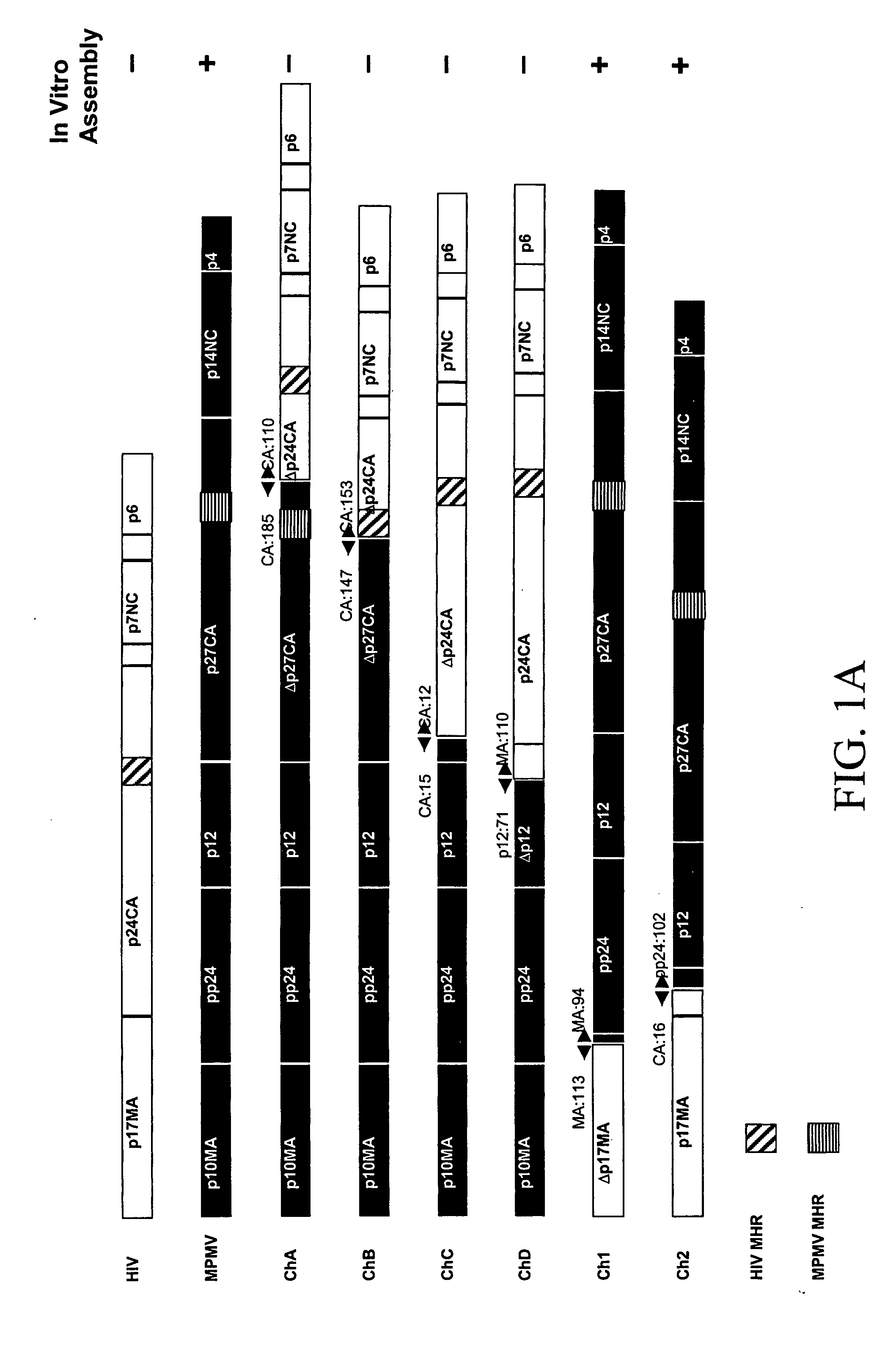
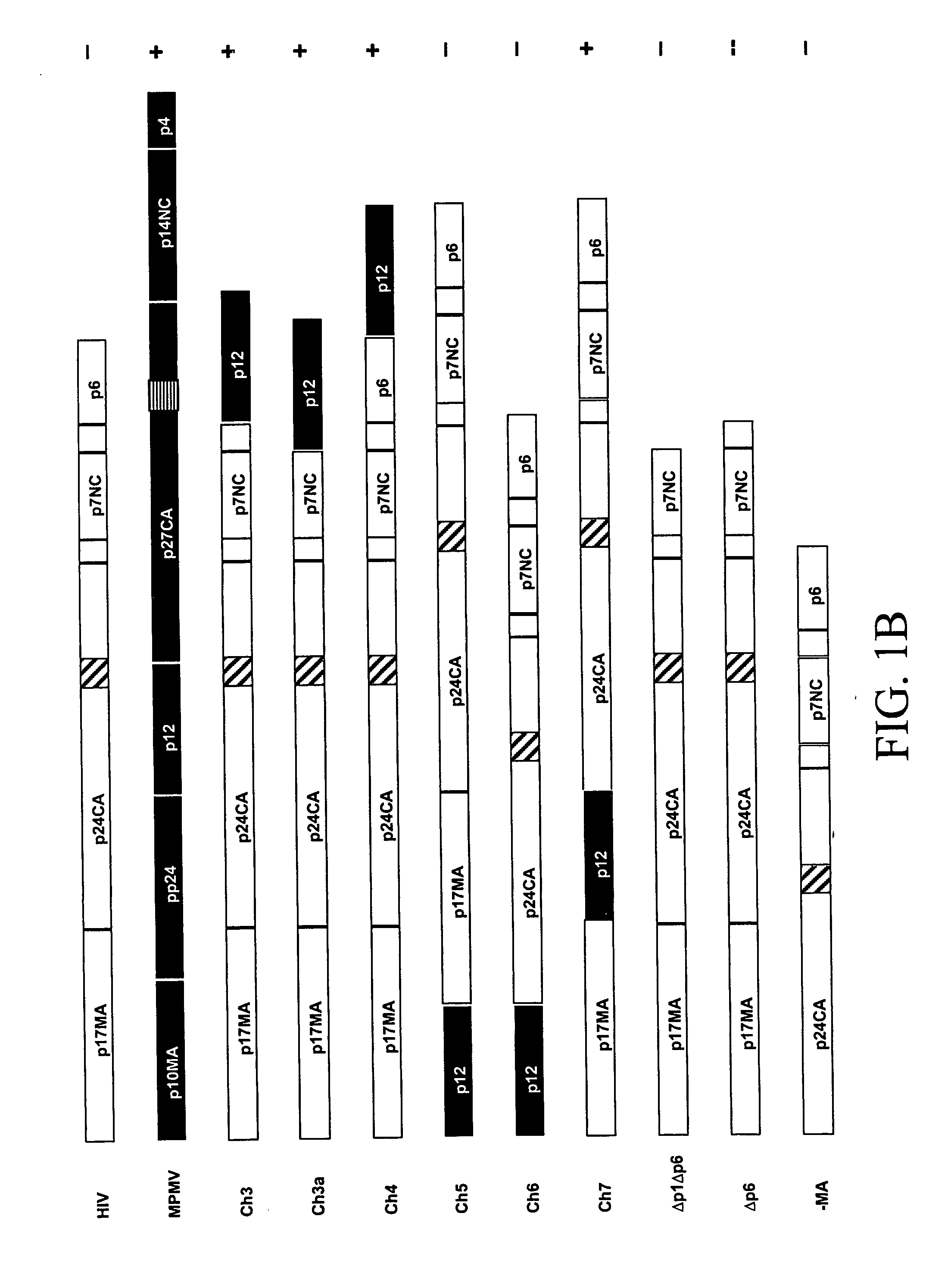
[0079] We have, surprisingly, found that the in vitro assembly of HIV capsids can be accomplished using chimeric M-PMV / HIV gag genes. A unique Gag domain of M-PMV (p12) is critical and required for chimeric Gag assembly in the in vitro translation system. We have found that the fusion of various portions of M-PMV Gag, including the p12 domain, with portions of HIV Gag, endows chimeric HIV Gag proteins with the ability to assemble in vitro.
[0080] We have constructed a number of chimeric gag genes, described below, with the purpose of endowing the HIV Gag precursor protein with the ability to assemble in our in vitro assay. As summarized in FIG. 1, the chimeric Gag polypeptides form assembled structures as assayed by gradient analysis.
[0081] Gag targets for association studies may also be constructed as a multivalent target protein. Bacterial expression vectors may be modified to contain an “Avitag” sequence in addition to the six-histidine tag. The “Avi...
example 2
Chimera Constructions
[0083] All DNA manipulations and cloning procedures were performed according to standard techniques. Before construction of the chimeric genes described below, it was first necessary to remove the ribosomal frameshift signal within the HIV gag coding sequence. Site-directed mutagenesis was employed to introduce a silent substitution in the “shifty sequence” of HIV gag. The loss of frameshift function was confirmed in an in vitro translation system.
[0084] The frameshift signal sequence in HIV gag is AAU UUU UUA GGG (SEQ ID NO:1). To introduce the silent mutations, PCR was performed with forward primer, GGCCAGATCTTCCCGAGGAAATTAGCCTG (SEQ ID NO:2), and reverse primer, ATAAGACAAGGACCAAAAG (SEQ ID NO:3). Plasmid pDAB72, obtained from the NIH AIDS Research and Reference Reagent Program, was used as the template. The PCR product was digested with Age I and Bgl II, and then ligated into similarly digested pDAB72. Clones were screened for the presence of an Ava I restr...
example 3
Binding Peptide Screening Assays
[0094] There are a number of assays by which peptide inhibitors of the subject invention may be identified. One such assay utilizes the yeast two-hybrid system for the identification of binding peptides. The binding peptides of this invention may reduce or inhibit the assembly of Gag polypeptides into capsids.
[0095] Chimeric Gag protein is expressed as a fusion with the GAL4-DNA-binding domain, and will be co-expressed with the GAL4 activation domain fused to a random sequence library encoding a decameric peptide or a semi-random peptide library. This approach has been used successfully to select peptides from a random library that could bind the Rb protein (Yang, et al. (1995)).
[0096] Construction of GAL-4-Gag Plasmids
[0097] The chimeric gag genes are cloned into GAL4 expression vectors well-known in the art (see for example, Wim Van Criekinge, et al. [1999]Biological Procedures Online, 2(1):1-38). For example, the complete chimeric gag gene from...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Tm | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com